Python 2.7です。
In [1]:
import sys
print(sys.version_info)
del sys
結合性解析の例¶
In [2]:
# Author: Martin Luessi <mluessi@nmr.mgh.harvard.edu>
#
# License: BSD (3-clause)
In [3]:
import numpy as np
import mne
mne.set_log_level('INFO')
from mne import io
from mne.connectivity import spectral_connectivity, seed_target_indices
from mne.datasets import sample
from mne.time_frequency import AverageTFR
print(__doc__) # doc stringというらしい 関数の説明らしいけど 深く考えないことにする。
パラメータの設定
In [4]:
data_path=sample.data_path()
raw_fname=data_path+'/MEG/sample/sample_audvis_filt-0-40_raw.fif'
event_fname=data_path+'/MEG/sample/sample_audvis_filt-0-40_raw-eve.fif'
# rawデータとeventの読み込み
raw=io.read_raw_fif(raw_fname)
events=mne.read_events(event_fname)
# 不良チャンネルの選択
raw.info['bads']+=['MEG 2443']
# gradiomterだけ選択
picks=mne.pick_types(
raw.info, meg='grad', eeg=False, stim=False, eog=True, exclude='bads')
# ecposhクラスの作成
event_id,tmin,tmax=3,-0.2,0.5
epochs=mne.Epochs(
raw,events,event_id,tmin,tmax,picks=picks,baseline=(None,0),
reject=dict(grad=4000e-13,eog=150e-6),preload=True)
# MEG 2343をseedとする
seed_ch='MEG 2343'
picks_ch_names=[raw.ch_names[i] for i in picks]
# seed-target indicesの計算
seed=picks_ch_names.index(seed_ch)
targets=np.arange(len(picks))
indices=seed_target_indices(seed,targets) # tuple 2 102と102
# waveletの定義
cwt_frequencies=np.arange(7,30,2)
cwt_n_cycles=cwt_frequencies/7.
# connectivityの計算 並列処理は2
sfreq=raw.info['sfreq']
con,freqs,times,_,_=spectral_connectivity( # conは204x11x106の三次元配列
epochs,indices=indices,method='wpli2_debiased',mode='cwt_morlet',sfreq=sfreq,
cwt_frequencies=cwt_frequencies,cwt_n_cycles=cwt_n_cycles,n_jobs=1)
In [5]:
# クラスepochsをfif形式で保存する場合
filename=data_path+'/MEG/sample/aaa-epo.fif'
epochs.save(filename)
# MATLABのMAT形式で保存する場合
import scipy.io
out_data={}
out_data['eopch']=epochs.get_data()
savename=data_path+'/MEG/sample/aaa.mat'
scipy.io.savemat(savename,out_data)
In [6]:
# 描画
# %matplotlib inline # いらないみたい
con[np.where(indices[1]==seed)]=1.0
title='WPLI2-Visual-Seed %s' % seed_ch
layout=mne.find_layout(epochs.info, 'meg')
tfr=AverageTFR(epochs.info,con, times,freqs, len(epochs))
print(type(tfr)) # クラス名表示
tfr.plot_topo(fig_facecolor='w',font_color='k',border='k');
ラベル指定した特定の信号源空間のconnectivity¶
In [7]:
% reset
In [8]:
# Authors: Alexandre Gramfort <alexandre.gramfort@telecom-paristech.fr>
#
# License: BSD (3-clause)
In [9]:
import matplotlib.pyplot as plt
import mne
mne.set_log_level('INFO')
from mne.datasets import sample
from mne.minimum_norm import apply_inverse_epochs, read_inverse_operator
from mne.connectivity import spectral_connectivity
print(__doc__)
data_path=sample.data_path()
subjects_dir=data_path+'/subjects'
fname_inv=data_path+'/MEG/sample/sample_audvis-meg-oct-6-meg-inv.fif'
fname_raw=data_path+'/MEG/sample/sample_audvis_filt-0-40_raw.fif'
fname_event=data_path+'/MEG/sample/sample_audvis_filt-0-40_raw-eve.fif'
# データ読み込み 空間フィルタ設計済み
inverse_operator=read_inverse_operator(fname_inv)
print(type(inverse_operator)) # クラスです
print(inverse_operator.keys()) # いろいろなクラスの集まりです
raw= mne.io.read_raw_fif(fname_raw)
events=mne.read_events(fname_event)
# 不良チャンネル
raw.info['bads']+=['MEG 2443']
# 脳磁図チャンネルと眼電図のみ利用
picks=mne.pick_types(raw.info,meg=True,eeg=False,stim=False,eog=True,exclude='bads')
# エポッククラスの作成
event_id,tmin,tmax=1,-0.2,0.5
epochs=mne.Epochs(
raw,events,event_id,tmin,tmax,picks=picks,baseline=(None,0),
reject=dict(mag=4e-12,grad=4000e-13,eog=150e-6))
# 信号源空間のエポックごとの計算
snr=1.0
lambda2=1.0/snr**2
method="dSPM" # MNE or sLORETAでもOK
# return_generator=Trueが大事らしい
stcs=apply_inverse_epochs(
epochs,inverse_operator,lambda2,method,pick_ori="normal",return_generator=True)
print(stcs) # generatorだそうです
# Label読み込み
names=['Aud-lh','Aud-rh','Vis-lh','Vis-rh']
labels=[mne.read_label(data_path+'/MEG/sample/labels/%s.label' % name)for name in names]
# 加算平均
src=inverse_operator['src'] # クラスSourceSpaces 要は三角メッシュ
label_ts=mne.extract_label_time_course(
stcs,labels,src,mode='mean_flip',return_generator=True)
print(label_ts) # generatorだそうです
fmin,fmax= 5.,40.
sfreq=raw.info['sfreq']
con,freqs,times,n_epochs,n_tapers=spectral_connectivity(
label_ts,method='wpli2_debiased',mode='multitaper',sfreq=sfreq,
fmin=fmin,fmax=fmax,mt_adaptive=True,n_jobs=1) # n_jobs=2だと遅くなる気がする
print(con.shape) # 4x4x25
n_rows,n_cols=con.shape[:2]
In [10]:
# 描画
fig,axes=plt.subplots(n_rows,n_cols,sharex=True,sharey=True)
plt.suptitle('Between labels connectivity')
for i in range(n_rows):
for j in range(i + 1):
if i==j:
axes[i,j].set_axis_off()
continue
axes[i,j].plot(freqs,con[i,j,:])
axes[j,i].plot(freqs,con[i,j,:])
if j==0:
axes[i,j].set_ylabel(names[i])
axes[0,i].set_title(names[i])
if i==(n_rows-1):
axes[i,j].set_xlabel(names[j])
axes[i,j].set_xlim([fmin,fmax])
axes[j,i].set_xlim([fmin,fmax])
# band limits
for f in [8,12,18,35]:
axes[i,j].axvline(f,color='k')
axes[j,i].axvline(f,color='k')
plt.show()
センサ空間(信号源)のconnectivity¶
In [11]:
% reset
In [12]:
# Author: Martin Luessi <mluessi@nmr.mgh.harvard.edu>
#
# License: BSD (3-clause)
In [13]:
import numpy as np
from scipy import linalg
import mne
mne.set_log_level('INFO')
from mne import io
from mne.connectivity import spectral_connectivity
from mne.datasets import sample
print(__doc__)
# ファイル名設定
data_path=sample.data_path()
raw_fname=data_path+'/MEG/sample/sample_audvis_filt-0-40_raw.fif'
event_fname=data_path+'/MEG/sample/sample_audvis_filt-0-40_raw-eve.fif'
# ファイル読み込み
raw=io.read_raw_fif(raw_fname)
events=mne.read_events(event_fname)
# 不良チャンネル
raw.info['bads']+=['MEG 2443']
# gradiometerと眼電図のみ選択
picks=mne.pick_types(raw.info,meg='grad',eeg=False,stim=False,eog=True,exclude='bads')
# エポッククラスの作成
event_id,tmin,tmax=3,-0.2,0.5
epochs=mne.Epochs(
raw,events,event_id,tmin,tmax,picks=picks,baseline=(None,0),
reject=dict(grad=4000e-13,eog=150e-6))
# 誘発応答を含んだまま結合性の計算
# 基線時間は省く
fmin,fmax=3.,9.
sfreq=raw.info['sfreq']
tmin=0.0
con,freqs,times,n_epochs,n_tapers=spectral_connectivity(
epochs, method='pli',mode='multitaper',sfreq=sfreq,fmin=fmin,fmax=fmax,
faverage=True,tmin=tmin,mt_adaptive=False,n_jobs=1)
print(con.shape) # 204x204x1
# 眼磁図雑音エポックを除去
ch_names=epochs.ch_names
idx=[ch_names.index(name) for name in ch_names if name.startswith('MEG')]
con=con[idx][:,idx]
# conは三次元配列
con = con[:, :, 0]
Mayaviを使います。Jupyterでは具合が悪いのでpython consoleを使います。
In [14]:
print(type(con))
print(con.shape)
filename=data_path+'/MEG/sample\\aaa.npz'# .npzはndarrayの拡張子 con.npyはダメみたい
np.savez(filename,con=con,idx=idx)
python consoleでは以下の図となります。
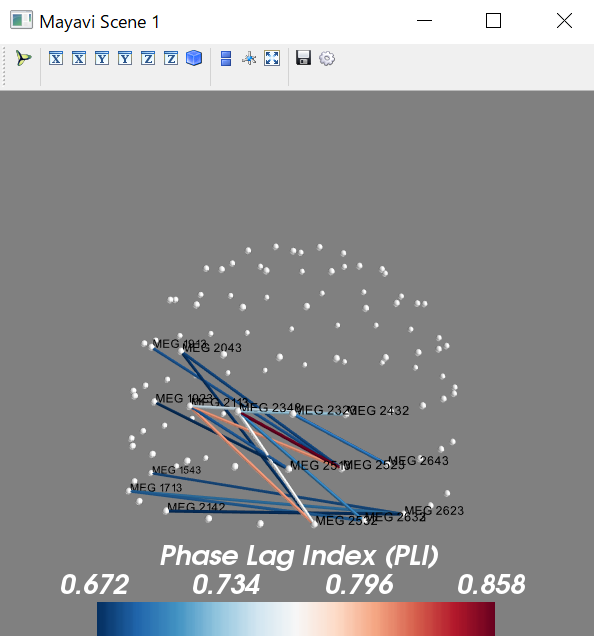
In [ ]:
# SpyderのConsoleで実行
import numpy as np
from scipy import linalg
import mne
from mne import io
from mne.datasets import sample
data_path=mne.datasets.sample.data_path()
filename=data_path+'/MEG/sample/aaa.npz'
x=np.load(filename)
con,idx=x['con'],x['idx']
raw_fname=data_path+'/MEG/sample/sample_audvis_filt-0-40_raw.fif'
raw=io.read_raw_fif(raw_fname)
picks=mne.pick_types(raw.info,meg='grad',eeg=False,stim=False,eog=True,exclude='bads')
from mayavi import mlab
mlab.figure(size=(600,600),bgcolor=(0.5,0.5,0.5));
sens_loc=[raw.info['chs'][picks[i]]['loc'][:3] for i in idx]
sens_loc=np.array(sens_loc)
pts=mlab.points3d(
sens_loc[:,0],sens_loc[:,1],sens_loc[:,2],color=(1,1,1),opacity=1,scale_factor=0.005);
n_con=20 # 最大20のconnection # connectionの最大値
min_dist=0.05 # 5cm以内のセンサは排除
threshold=np.sort(con,axis=None)[-n_con]
ii,jj=np.where(con>=threshold)
con_nodes=list()
con_val=list()
for i, j in zip(ii,jj):
if linalg.norm(sens_loc[i]-sens_loc[j])>min_dist:
con_nodes.append((i,j))
con_val.append(con[i,j])
con_val=np.array(con_val)
vmax=np.max(con_val)# 描画
vmin=np.min(con_val)
for val,nodes in zip(con_val,con_nodes):
x1,y1,z1=sens_loc[nodes[0]]
x2,y2,z2=sens_loc[nodes[1]]
points=mlab.plot3d(
[x1,x2],[y1,y2],[z1,z2],[val,val],vmin=vmin,vmax=vmax,
tube_radius=0.001,colormap='RdBu');
points.module_manager.scalar_lut_manager.reverse_lut = True
mlab.scalarbar(title='Phase Lag Index (PLI)',nb_labels=4)
nodes_shown=list(set([n[0] for n in con_nodes]+[n[1] for n in con_nodes]))
for node in nodes_shown:
x,y,z=sens_loc[node]
mlab.text3d(x,y,z,raw.ch_names[picks[node]],scale=0.005,color=(0,0,0));
view=(-88.7,40.8,0.76, np.array([-3.9e-4,-8.5e-3,-1e-2]))
mlab.view(*view)
視覚刺激時の信号源空間のPhase Slope Index (PSI)の計算¶
In [15]:
% reset
In [16]:
# Author: Martin Luessi <mluessi@nmr.mgh.harvard.edu>
#
# License: BSD (3-clause)
IPython環境になるようで、ちょっと時間がかかります。
In [17]:
import numpy as np
import mne
from mne.datasets import sample
from mne.minimum_norm import read_inverse_operator, apply_inverse_epochs
from mne.connectivity import seed_target_indices, phase_slope_index
print(__doc__)
In [18]:
data_path=sample.data_path()
subjects_dir=data_path+'/subjects'
fname_inv=data_path+'/MEG/sample/sample_audvis-meg-oct-6-meg-inv.fif' # 計算済み
fname_raw=data_path+'/MEG/sample/sample_audvis_filt-0-40_raw.fif'
fname_event=data_path+'/MEG/sample/sample_audvis_filt-0-40_raw-eve.fif'
fname_label=data_path+'/MEG/sample/labels/Vis-lh.label'
event_id,tmin,tmax= 4,-0.2,0.3
method="dSPM" # use dSPM method (could also be MNE or sLORETA)
# データ読み込み
inverse_operator=read_inverse_operator(fname_inv)
print(type(inverse_operator)) # クラスです
print(inverse_operator.keys()) # いろいろなクラスの集まりです
raw=mne.io.read_raw_fif(fname_raw)
events=mne.read_events(fname_event)
picks=mne.pick_types(raw.info,meg=True,eeg=False,stim=False,eog=True,exclude='bads')
# エポッククラスの作成
epochs=mne.Epochs(
raw,events,event_id,tmin, tmax,picks=picks,baseline=(None, 0),
reject=dict(mag=4e-12,grad=4000e-13,eog=150e-6))
# 空間フィルタの作成
snr=1.0
lambda2=1.0/snr**2
stcs=apply_inverse_epochs( # generatorだそうです return_generator=Falseは数が少ないから?
epochs,inverse_operator,lambda2,method,pick_ori="normal",return_generator=False)
# seed time seriesの計算
label=mne.read_label(fname_label) #Vis-lh.label
src=inverse_operator['src'] # 信号源空間を利用
seed_ts=mne.extract_label_time_course(stcs,label,src,mode='mean_flip')
# seed time courseと信号源推定
# index 0: Vis-lh.labelの経時変化
# index 1..7499: 信号源空間の経時変化
comb_ts=zip(seed_ts,stcs)
# connectivityの計算
vertices=[src[i]['vertno'] for i in range(2)]
n_signals_tot=1+len(vertices[0])+len(vertices[1])
indices=seed_target_indices([0],np.arange(1,n_signals_tot))
# 8~30HzでPhase Slow Index baseline期間は含まず
fmin,fmax,tmin_con=8.,30.,0.
sfreq=raw.info['sfreq']
psi,freqs,times,n_epochs,_=phase_slope_index(
comb_ts,mode='multitaper',indices=indices,sfreq=sfreq,fmin=fmin,fmax=fmax,tmin=tmin_con)
psi_stc=mne.SourceEstimate(psi,vertices=vertices,tmin=0,tstep=1,subject='sample')
In [19]:
# データ保存
fname_psi=fname_raw=data_path+'/MEG/sample/psi.npy'
fname_psi_stc=fname_raw=data_path+'/MEG/sample/psi.stc'
np.save(fname_psi,psi)
psi_stc.save(fname_psi_stc)
In [20]:
# python console側で読みだすファイル名
print(fname_psi)
print(fname_psi_stc)
print(subjects_dir)
print(fname_label)
3DなのでPython Consoleを使います。
以下のような図が描画されます。
python consoleでは以下の図となります。
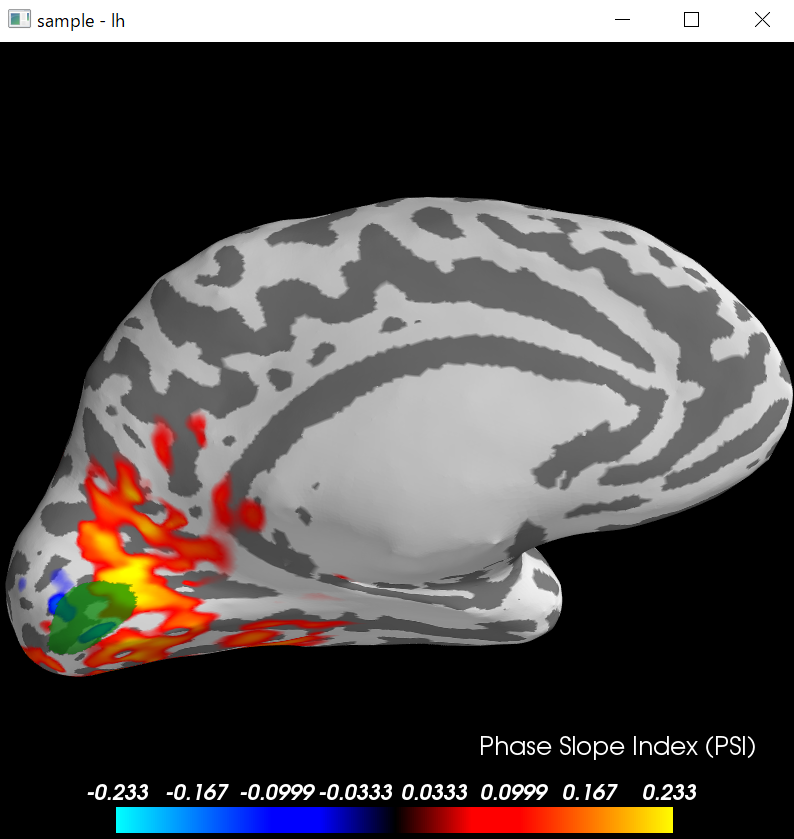

In [ ]:
# SpyderのPython Consoleで実行
import numpy as np
import mne
data_path=mne.datasets.sample.data_path()
psi=np.load(data_path+'/MEG/sample/psi.npy')
psi_stc=mne.read_source_estimate(data_path+'/MEG/sample/psi.stc')
psi_stc.subject='sample' # これは何故か保存されてないみたい
subjects_dir=data_path+'/subjects'
v_max=np.max(np.abs(psi))
brain=psi_stc.plot(
surface='inflated',hemi='lh',time_label='Phase Slope Index (PSI)',
subjects_dir=subjects_dir,clim=dict(kind='percent',pos_lims=(95, 97.5, 100)))
brain.show_view('medial')
fname_label=data_path+'/MEG/sample/labels/Vis-lh.label'
brain.add_label(fname_label,color='green',alpha=0.7)
信号源空間でのcoherence¶
In [21]:
% reset
In [22]:
# Author: Martin Luessi <mluessi@nmr.mgh.harvard.edu>
#
# License: BSD (3-clause)
In [23]:
import numpy as np
import mne
from mne.datasets import sample
from mne.minimum_norm import (apply_inverse, apply_inverse_epochs,read_inverse_operator)
from mne.connectivity import seed_target_indices, spectral_connectivity
print(__doc__)
data_path=sample.data_path()
subjects_dir=data_path + '/subjects'
fname_inv=data_path + '/MEG/sample/sample_audvis-meg-oct-6-meg-inv.fif'
fname_raw=data_path + '/MEG/sample/sample_audvis_filt-0-40_raw.fif'
fname_event=data_path + '/MEG/sample/sample_audvis_filt-0-40_raw-eve.fif'
label_name_lh='Aud-lh'
fname_label_lh=data_path + '/MEG/sample/labels/%s.label' % label_name_lh
event_id,tmin,tmax=1,-0.2,0.5
method="dSPM"
In [24]:
inverse_operator=read_inverse_operator(fname_inv)
label_lh=mne.read_label(fname_label_lh) # 左聴覚野をseedにする
raw=mne.io.read_raw_fif(fname_raw)
events=mne.read_events(fname_event)
raw.info['bads']+=['MEG 2443']
picks=mne.pick_types(raw.info,meg=True,eeg=False,stim=False,eog=True,exclude='bads')
epochs = mne.Epochs(
raw,events,event_id,tmin,tmax,picks=picks,baseline=(None,0),
reject=dict(mag=4e-12,grad=4000e-13,eog=150e-6))
snr=3.0
lambda2=1.0/snr**2
evoked=epochs.average()
stc=apply_inverse(evoked,inverse_operator,lambda2,method,pick_ori="normal")
stc_label=stc.in_label(label_lh) # 左半球に限定
src_pow=np.sum(stc_label.data**2,axis=1)
seed_vertno=stc_label.vertices[0][np.argmax(src_pow)]
seed_idx=np.searchsorted(stc.vertices[0],seed_vertno)
n_sources=stc.data.shape[0]
indices=seed_target_indices([seed_idx],np.arange(n_sources))
# return_generator=Trueにすると信号源データなしで計算できてメモリ節約できる
snr=1.0
lambda2=1.0/snr**2
stcs=apply_inverse_epochs(
epochs,inverse_operator,lambda2,method,pick_ori="normal",return_generator=True)
print(stcs) # generatorだそうです
fmin=(8.,13.) # Hz
fmax=(13.,30.) # Hz
sfreq=raw.info['sfreq']
coh,freqs,times,n_epochs,n_tapers=spectral_connectivity(
stcs,method='coh',mode='fourier',indices=indices,sfreq=sfreq,fmin=fmin,
fmax=fmax,faverage=True,n_jobs=1)
print('Frequencies in Hz over which coherence was averaged for alpha: ')
print(freqs[0])
print('Frequencies in Hz over which coherence was averaged for beta: ')
print(freqs[1])
# seed1個で信号源のconnectivityの計算
tmin=np.mean(freqs[0])
tstep=np.mean(freqs[1])-tmin
coh_stc= mne.SourceEstimate(
coh,vertices=stc.vertices,tmin=1e-3*tmin,tstep=1e-3*tstep,subject='sample')
In [25]:
stcs
Out[25]:
In [26]:
# データ保存
fname_coh_stc=fname_raw=data_path+'/MEG/sample/coh.stc'
coh_stc.save(fname_coh_stc)
SpyderのConsoleで実行すると以下の図となります。
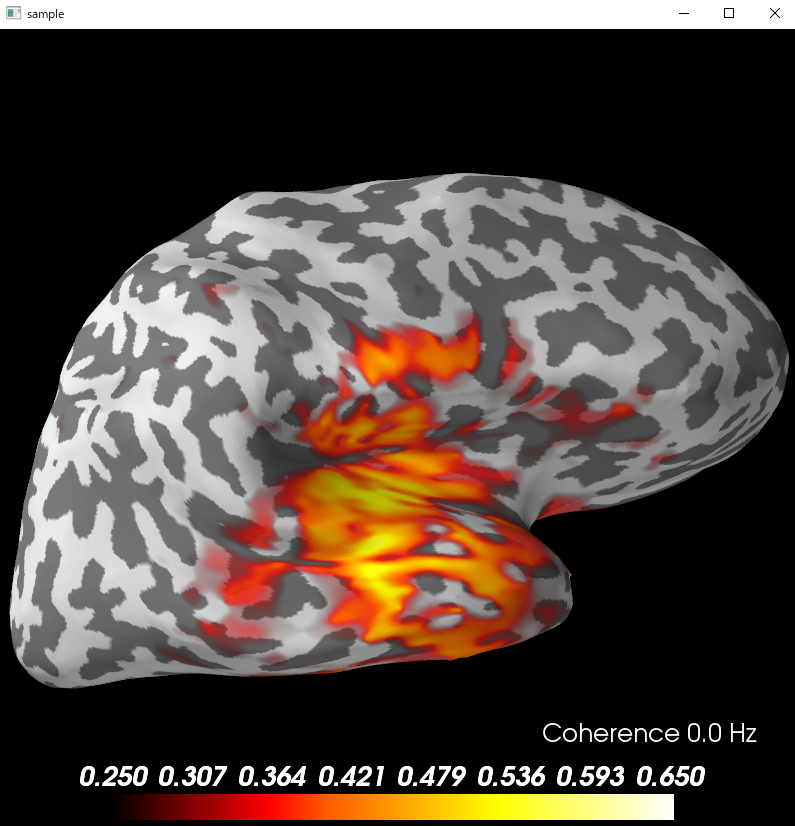
In [ ]:
# SpyderのPython Consoleで実行
import mne
data_path=mne.datasets.sample.data_path()
coh_stc=mne.read_source_estimate(data_path+'/MEG/sample/coh.stc')
coh_stc.subject='sample' # これは何故か保存されてないみたい
subjects_dir=data_path+'/subjects'
brain=coh_stc.plot('sample','inflated','both',time_label='Coherence %0.1f Hz',subjects_dir=subjects_dir,
clim=dict(kind='value',lims=(0.25, 0.4, 0.65)))
brain.show_view('lateral')
信号源空間の結合性の円形表示¶
In [27]:
% reset
In [28]:
# Authors: Martin Luessi <mluessi@nmr.mgh.harvard.edu>
# Alexandre Gramfort <alexandre.gramfort@telecom-paristech.fr>
# Nicolas P. Rougier (graph code borrowed from his matplotlib gallery)
#
# License: BSD (3-clause)
In [29]:
import mne
from mne.datasets import sample
from mne.minimum_norm import apply_inverse_epochs,read_inverse_operator
from mne.connectivity import spectral_connectivity
from mne.viz import circular_layout,plot_connectivity_circle
print(__doc__)
data_path=sample.data_path()
subjects_dir=data_path+'/subjects'
fname_inv=data_path+'/MEG/sample/sample_audvis-meg-oct-6-meg-inv.fif'
fname_raw=data_path+'/MEG/sample/sample_audvis_filt-0-40_raw.fif'
fname_event=data_path+'/MEG/sample/sample_audvis_filt-0-40_raw-eve.fif'
inverse_operator=read_inverse_operator(fname_inv) # 計算済み
print(type(inverse_operator)) # クラスです
print(inverse_operator.keys()) # いろいろなクラスの集まりです
raw=mne.io.read_raw_fif(fname_raw)
events= mne.read_events(fname_event) # 処理済み
raw.info['bads']+=['MEG 2443']
picks=mne.pick_types(raw.info,meg=True,eeg=False,stim=False,eog=True,exclude='bads')
# クラスepochの作成
event_id,tmin,tmax=1,-0.2,0.5
epochs=mne.Epochs(
raw,events,event_id,tmin,tmax,picks=picks,baseline=(None,0),
reject=dict(mag=4e-12,grad=4000e-13,eog=150e-6))
# 信号源空間dSPMの計算 MNE, sLORETAも可
snr=1.0
lambda2=1.0/snr**2
stcs=apply_inverse_epochs(
epochs,inverse_operator,lambda2,'dSPM',pick_ori="normal",return_generator=True)
print(stcs) # generatorだそうです
# FreeSurferのLabel取得
labels=mne.read_labels_from_annot('sample',parc='aparc',subjects_dir=subjects_dir)
label_colors=[label.color for label in labels]
print(len(labels)) # 68個のラベルです。
# return_generator=Trueが定番です。
src=inverse_operator['src'] #SourceSpacesクラス 要するにメッシュ
label_ts=mne.extract_label_time_course(
stcs,labels,src,mode='mean_flip',return_generator=True)
print(label_ts) # generatorだそうです。
α帯域の結合性の計算¶
1) rawからepochを1つ読み込む
2) SSP適応と基線補正
3) 信号源波形の計算
4) 各部位(label)毎に信号源波形の加算平
5) 結合性の計算し、1)次のepoch
epoch毎に計算することでメモリ消費を抑える
In [30]:
# 結合性の計算
import numpy as np
import matplotlib.pyplot as plt
# % matplotlib inline # いらないみたい
fmin,fmax=8.,13.
sfreq=raw.info['sfreq']
con_methods=['pli','wpli2_debiased']
con,freqs,times,n_epochs,n_tapers=spectral_connectivity( # ここで改行すると4文字タブとなる
label_ts,method=con_methods,mode='multitaper',sfreq=sfreq,fmin=fmin,
fmax=fmax,faverage=True,mt_adaptive=True,n_jobs=1)
# conは三次元配列のリスト
print(con[0].shape)
print(con[1].shape)
con_res= dict()
for method, c in zip(con_methods,con):
con_res[method]=c[:,:,0]
# 描画
label_names=[label.name for label in labels]
lh_labels=[name for name in label_names if name.endswith('lh')] # 左半球
label_ypos=list()
for name in lh_labels:
idx=label_names.index(name)
ypos=np.mean(labels[idx].pos[:,1])
label_ypos.append(ypos)
lh_labels=[label for (yp,label) in sorted(zip(label_ypos, lh_labels))]
rh_labels=[label[:-2]+'rh' for label in lh_labels] # 右半球
node_order=list()
node_order.extend(lh_labels[::-1]) # reverse the order
node_order.extend(rh_labels)
node_angles=circular_layout(
label_names,node_order,start_pos=90,group_boundaries=[0,len(label_names)/2])
# 上位300のみ表示
plot_connectivity_circle(
con_res['pli'],label_names,n_lines=300,node_angles=node_angles,node_colors=label_colors,
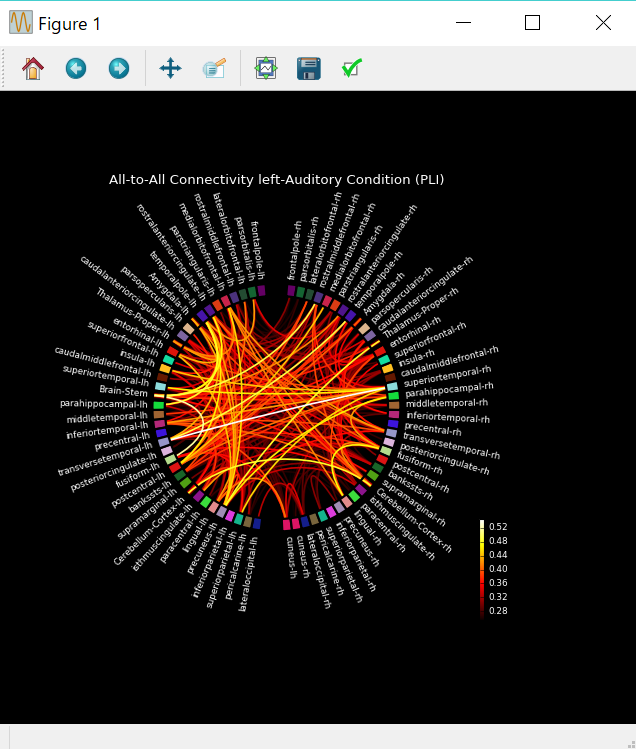
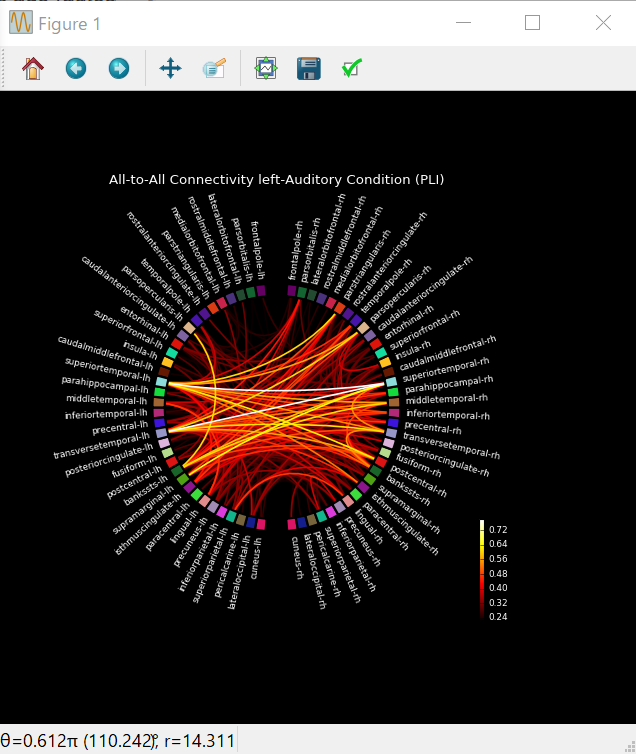
title='All-to-All Connectivity left-Auditory ' 'Condition (PLI)') # 'A' 'B' = 'AB'
plt.savefig('circle.png',facecolor='black')
# なんかうまく表示されません
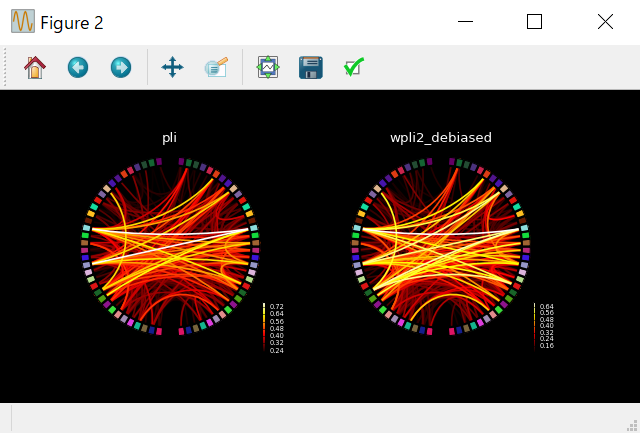
fig=plt.figure(num=None,figsize=(8,4),facecolor='black')
no_names=['']*len(label_names)
for ii, method in enumerate(con_methods):
plot_connectivity_circle(
con_res[method],no_names,n_lines=300,node_angles=node_angles,node_colors=label_colors,
title=method,padding=0,fontsize_colorbar=6,fig=fig,subplot=(1,2,ii+1))
plt.show()
pickleを使ってpythonのオブジェクトデータをファイル保存¶
これとっても便利です。但し関数とかバイトデータになってないものは保存できません。クラスをそのまま保存しようとしてもダメだそうです。
In [31]:
# データを保存します。
import pickle
filename=data_path+'/MEG/sample/con_res.pkl'
with open(filename,mode='wb') as f: # withを使うとf.close()が不要
pickle.dump(con_res,f)
filename=data_path+'/MEG/sample/label_names.pkl'
with open(filename,mode='wb') as f:
pickle.dump(label_names,f)
filename=data_path+'/MEG/sample/node_angles.pkl'
with open(filename,mode='wb') as f:
pickle.dump(node_angles,f)
filename=data_path+'/MEG/sample/label_colors.pkl'
with open(filename,mode='wb') as f:
pickle.dump(label_colors,f)
filename=data_path+'/MEG/sample/con_methods.pkl'
with open(filename,mode='wb') as f:
pickle.dump(con_methods,f)
Python Consoleだと図は正しく表示されました。
In [ ]:
# SpyderのPython Consoleで実行
import matplotlib.pyplot as plt
import pickle
import mne
data_path=mne.datasets.sample.data_path()
from mne.viz import plot_connectivity_circle
pathname=data_path+'/MEG/sample/'
filename=pathname+'con_res.pkl'
with open(filename,mode='rb') as f:
con_res=pickle.load(f);
filename=pathname+'label_names.pkl'
with open(filename,mode='rb') as f:
label_names=pickle.load(f);
filename=pathname+'node_angles.pkl'
with open(filename,mode='rb') as f:
node_angles=pickle.load(f);
filename=pathname+'label_colors.pkl'
with open(filename,mode='rb') as f:
label_colors=pickle.load(f);
filename=pathname+'con_methods.pkl'
with open(filename,mode='rb') as f:
con_methods=pickle.load(f);
plot_connectivity_circle(
con_res['pli'],label_names,n_lines=300,node_angles=node_angles,node_colors=label_colors,
title='All-to-All Connectivity left-Auditory ' 'Condition (PLI)') # 'A' 'B' = 'AB'
plt.savefig('circle.png',facecolor='black')
fig=plt.figure(num=None,figsize=(8,4),facecolor='black')
no_names=['']*len(label_names)
for ii, method in enumerate(con_methods):
plot_connectivity_circle(
con_res[method],no_names,n_lines=300,node_angles=node_angles,node_colors=label_colors,
title=method,padding=0,fontsize_colorbar=6,fig=fig,subplot=(1,2,ii+1))
plt.show()
信号源空間の結合性の円形表示¶
In [32]:
# Author: Annalisa Pascarella <a.pascarella@iac.cnr.it>
#
# License: BSD (3-clause)
In [33]:
% reset
予めFreeSurferColorLUT.txtを所定のフォルダに保存しておく必要があります。FreeSurferColorLUT.txtはLinuxの場合、/sur/local/freesurferにあります。ネットで検索したものでもOKです。保存先はC:/Users/username/Anaconda2/lib/site-packages/mne/dataです。
In [34]:
import os.path as op
import numpy as np
import mne
from mne.datasets import sample
from mne import setup_volume_source_space,setup_source_space
from mne import make_forward_solution
from mne.io import read_raw_fif
from mne.minimum_norm import make_inverse_operator,apply_inverse_epochs
from mne.connectivity import spectral_connectivity
from mne.viz import circular_layout,plot_connectivity_circle
data_path=sample.data_path()
subject='sample'
data_dir=op.join(data_path,'MEG',subject)
subjects_dir=op.join(data_path,'subjects')
bem_dir=op.join(subjects_dir,subject,'bem')
fname_aseg=op.join(subjects_dir,subject,'mri','aseg.mgz')
fname_model=op.join(bem_dir,'%s-5120-bem.fif' % subject)
fname_bem=op.join(bem_dir,'%s-5120-bem-sol.fif' % subject)
fname_raw=data_dir+'/sample_audvis_filt-0-40_raw.fif'
fname_trans=data_dir+'/sample_audvis_raw-trans.fif'
fname_cov=data_dir+'/ernoise-cov.fif'
fname_event=data_dir+'/sample_audvis_filt-0-40_raw-eve.fif'
labels_vol=[
'Left-Amygdala','Left-Thalamus-Proper','Left-Cerebellum-Cortex','Brain-Stem',
'Right-Amygdala','Right-Thalamus-Proper', 'Right-Cerebellum-Cortex']
src=setup_source_space(
subject,fname=None,subjects_dir=subjects_dir,spacing='oct6',add_dist=False)
vol_src=setup_volume_source_space(
subject,mri=fname_aseg,pos=7.0,bem=fname_model,
volume_label=labels_vol,subjects_dir=subjects_dir)
src+=vol_src
fwd=make_forward_solution(
fname_raw,fname_trans,src,fname_bem,mindist=5.0, # innerskullから5mm以内は無視
meg=True,eeg=False,n_jobs=1)
raw=read_raw_fif(fname_raw,preload=True)
noise_cov=mne.read_cov(fname_cov)
events=mne.read_events(fname_event)
raw.info['bads']+=['MEG 2443']
picks=mne.pick_types(raw.info,meg=True,eeg=False,stim=False,eog=True,exclude='bads')
event_id,tmin,tmax=1,-0.2,0.5
epochs=mne.Epochs(
raw,events,event_id,tmin,tmax,picks=picks,baseline=(None,0),
reject=dict(mag=4e-12,grad=4000e-13,eog=150e-6))
snr=1.0
inv_method='dSPM'
parc='aparc'
lambda2=1.0/snr**2
inverse_operator=make_inverse_operator(
raw.info,fwd,noise_cov,loose=None,depth=None,fixed=False)
stcs=apply_inverse_epochs(
epochs,inverse_operator,lambda2,inv_method,pick_ori=None,return_generator=True)
labels_parc = mne.read_labels_from_annot(subject, parc=parc,
subjects_dir=subjects_dir)
src=inverse_operator['src']
label_ts=mne.extract_label_time_course(
stcs,labels_parc,src,mode='mean_flip',allow_empty=True,return_generator=False)
fmin,fmax=8.,13.
sfreq=raw.info['sfreq']
con,freqs,times,n_epochs,n_tapers=spectral_connectivity(
label_ts,method='pli',mode='multitaper',sfreq=sfreq,fmin=fmin,
fmax=fmax,faverage=True,mt_adaptive=True,n_jobs=1)
labels_aseg=mne.get_volume_labels_from_src(src,subject,subjects_dir)
labels=labels_parc+labels_aseg
node_colors=[label.color for label in labels]
label_names=[label.name for label in labels]
lh_labels=[name for name in label_names if name.endswith('lh')]
rh_labels=[name for name in label_names if name.endswith('rh')]
label_ypos_lh=list()
for name in lh_labels:
idx=label_names.index(name)
ypos=np.mean(labels[idx].pos[:,1])
label_ypos_lh.append(ypos)
try:
idx=label_names.index('Brain-Stem')
ypos=np.mean(labels[idx].pos[:,1])
lh_labels.append('Brain-Stem')
label_ypos_lh.append(ypos)
except ValueError:
pass
lh_labels=[label for (yp,label) in sorted(zip(label_ypos_lh,lh_labels))]
rh_labels=[label[:-2]+'rh' for label in lh_labels
if label!='Brain-Stem' and label[:-2]+'rh' in rh_labels]
node_order=list()
node_order=lh_labels[::-1]+rh_labels
node_angles=circular_layout(
label_names,node_order, start_pos=90,group_boundaries=[0,len(label_names)//2])
conmat=con[:,:,0]
In [35]:
# データを保存します。
import pickle
pathname=data_path+'/MEG/sample/'
filename=pathname+'conmat.pkl' # conは予約語ではないが、ファイル名としては保存されないっぽい。
with open(filename,mode='wb') as f: # withを使うとf.close()が不要
pickle.dump(conmat,f)
filename=pathname+'label_names.pkl'
with open(filename,mode='wb') as f:
pickle.dump(label_names,f)
filename=pathname+'node_angles.pkl'
with open(filename,mode='wb') as f:
pickle.dump(node_angles,f)
filename=pathname+'node_colors.pkl'
with open(filename,mode='wb') as f:
pickle.dump(node_colors,f)
In [ ]:
# SpyderのPython Consoleで実行
import matplotlib.pyplot as plt
import pickle
import mne
data_path=mne.datasets.sample.data_path()
from mne.viz import plot_connectivity_circle
pathname=data_path+'/MEG/sample/'
filename=pathname+'conmat.pkl'
with open(filename,mode='rb') as f:
conmat=pickle.load(f)
filename=pathname+'label_names.pkl'
with open(filename,mode='rb') as f:
label_names=pickle.load(f);
filename=pathname+'node_angles.pkl'
with open(filename,mode='rb') as f:
node_angles=pickle.load(f);
filename=pathname+'node_colors.pkl'
with open(filename,mode='rb') as f:
node_colors=pickle.load(f);
plot_connectivity_circle(
conmat,label_names,n_lines=300,node_angles=node_angles,node_colors=node_colors,
title='All-to-All Connectivity left-Auditory Condition (PLI)');
'''
import matplotlib.pyplot as plt
plt.savefig('circle.png', facecolor='black')
'''