BrainVISAの結果をMNE-Cで使う方法
MNE-Cではvolume dataであるT1.mgz、brain.mgz、polygon mesh dataであるlh.white、rh.whiteがあればとりあえずdSPMで電流源の推定結果を表示することは可能です。
BrainVISAでは
~/default_acuisition/subject.nii.gz、(/mri/T1.mgz)
~/default_acquisition/default_analysis/segmentation/brain_subject.nii.gz、(/mri/brain.mgz 但しT1.mgzデータのbrain.mgz部分)
~/default_acuisition/default_analysis/segmentation/mesh/subject_Lwhite.gi、(/surf/lh.white)
~/default_acquisition/default_analysis/segmentation/mesh/subject_Rwhite.gi、(/surf/rh.white)
がそれに相当します。
NIfTIファイルからmgzファイルへの変換はFreeSurferのmri_convet、
GIfTIファイルからsurfファイルへの変換はFreeSurferのmris_convert
で行います。が、座標系の変換はしてくれませんので書き換える必要があります。
MATLAB R2017b以降でNIfTIファイルの読み書きが可能になっています。またGIfTIはGIfTI toolbox for MATLABを使うことでファイルの読み書きが可能です。
MNE MATLAB toolboxというのがあるのはあるのですが、基本的にFIFFファイル読み書き用のtoolです。FreeSurferのsurfファイルは読み書きできるものの、MGZファイルは扱えないようです。
座標変換がない場合
水平断のDICOM画像をNIfTI化し、BrainVISAで処理したNIfTIデータとFreeSurferで処理したMGZデータを比較します。
MATLAB R2018aのコードです
尚、MGZファイルはFreeSurferのmri_convertコマンドでNIfTIファイル化しています。
clear;
close all;
a.T1.data=niftiread('c:\test\a\mri\T1.nii.gz');
a.brain.data=niftiread('c:\test\a\mri\brain.nii.gz');
% a.T1.info=niftiinfo('c:\test\a\mri\brain.nii.gz');
% 読み込みエラーになる
a.brain.info=niftiinfo('c:\test\a\mri\brain.nii.gz');
b.T1.data=niftiread('c:\test\b\mri\T1.nii.gz');
b.brain.info=niftiinfo('c:\test\b\mri\brain.nii.gz');
figure('color',[1,1,1]);
colormap(gray(256));
for n=1:2
if n==1
Z=a.T1.data;
scale=a.brain.info.PixelDimensions;
else
Z=b.T1.data;
scale=b.brain.info.PixelDimensions;
end
subplot(2,3,n*3-2);
imagesc(squeeze(Z(size(Z,1)/2,:,:)));
daspect([scale(2),scale(3),scale(1)]);
subplot(2,3,n*3-1);
imagesc(squeeze(Z(:,size(Z,2)/2,:)));
daspect([scale(1),scale(3),scale(2)]);
subplot(2,3,n*3);
imagesc(squeeze(Z(:,:,size(Z,3)/2)));
daspect([scale(1),scale(2),scale(3)]);
end
set(findobj(gcf,'type','axes'),'clim',[0,255]);
上段がBrainVISA、下段がFreeSurferです。三次元配列の第2・3軸、第1・3軸、第1・2軸の順となっています。
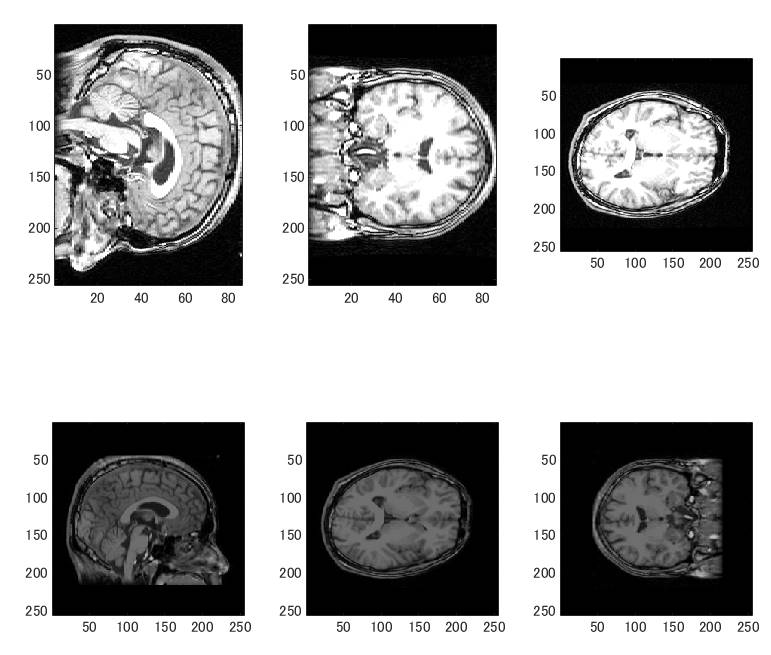
BrinVISAでは座標系は右左、後前、下上の順になっています。
FreeSurferでは座標系は右左、上下、後前の順になっています。
これらの画像をもとに脳を切り出してpolygon mesh化したデータを比較します。
尚surfファイルはmris_convertコマンドで予めGIfTIファイルに変換しておきます。
a.meshL=gifti('c:\test\a\surf\Lwhite.gii');
a.meshR=gifti('c:\test\a\surf\Rwhite.gii');
% 何故かmris_convert lh.white Lwhite.giiはできず
b.meshL=gifti('c:\test\b\surf\lh.white.gii');
b.meshR=gifti('c:\test\b\surf\rh.white.gii');
figure('color',[1,1,1]);
for n=1:2
if n==1
p1=a.meshL.vertices;
t1=a.meshL.faces;
p2=a.meshR.vertices;
t2=a.meshR.faces;
else
p1=b.meshL.vertices;
t1=b.meshL.faces;
p2=b.meshR.vertices;
t2=b.meshR.faces;
end
subplot(2,3,n*3-2);
meshL=trimesh(t1,p1(:,1),p1(:,2),p1(:,3),'LineStyle','none','facecolor',[0,0,1],'facealpha',0.1);hold
on;
meshR=trimesh(t2,p2(:,1),p2(:,2),p2(:,3),'LineStyle','none','facecolor',[0,1,0],'facealpha',0.1);
view([0,0]);daspect([1,1,1]);axis tight;grid on;
subplot(2,3,n*3-1);
copyobj(meshL,gca);hold on;
copyobj(meshR,gca);
view([90,0]);daspect([1,1,1]);axis tight;grid on;
subplot(2,3,n*3);
copyobj(meshL,gca);hold on;
copyobj(meshR,gca);
view([0,90]);daspect([1,1,1]);axis tight;grid on;
end
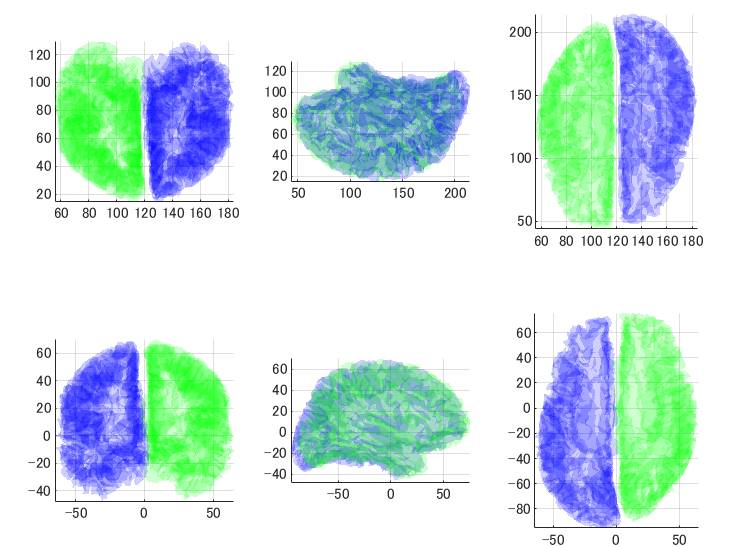
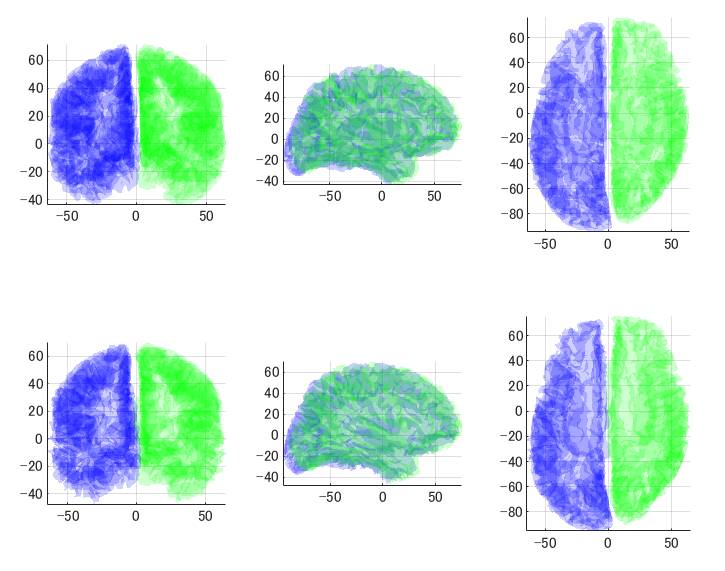
上がBrainVISA、下がFreeSurferです。青が左半球、緑が右半球です。左が正面、中央が左側面、右が上から見たところです。
BrainVISAは中点が128くらい、FreeSurferは中点は0くらいになっています。
MNE-CはFreeSurfer用の座標でないとうまく電流源推定できません。
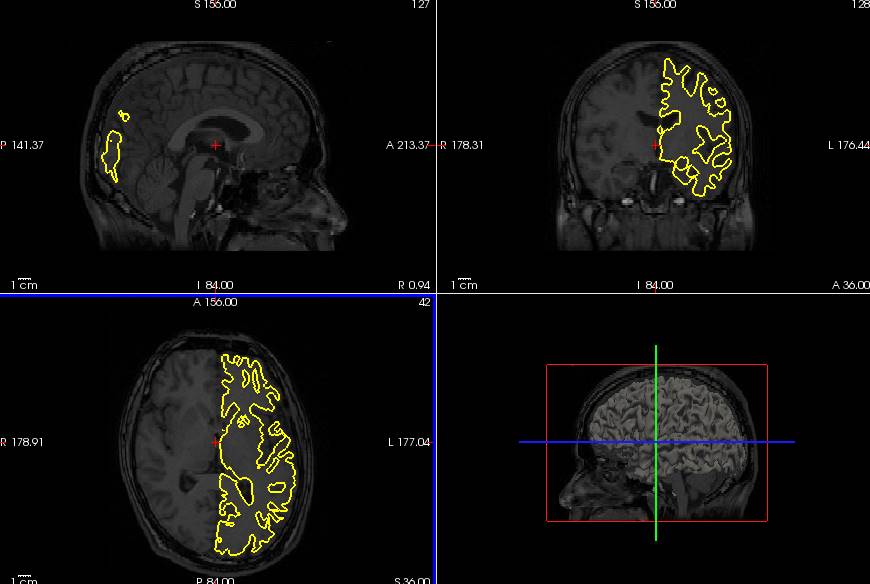
freeviewでT1.ni.gzとLwhite.giiを開いてみました。
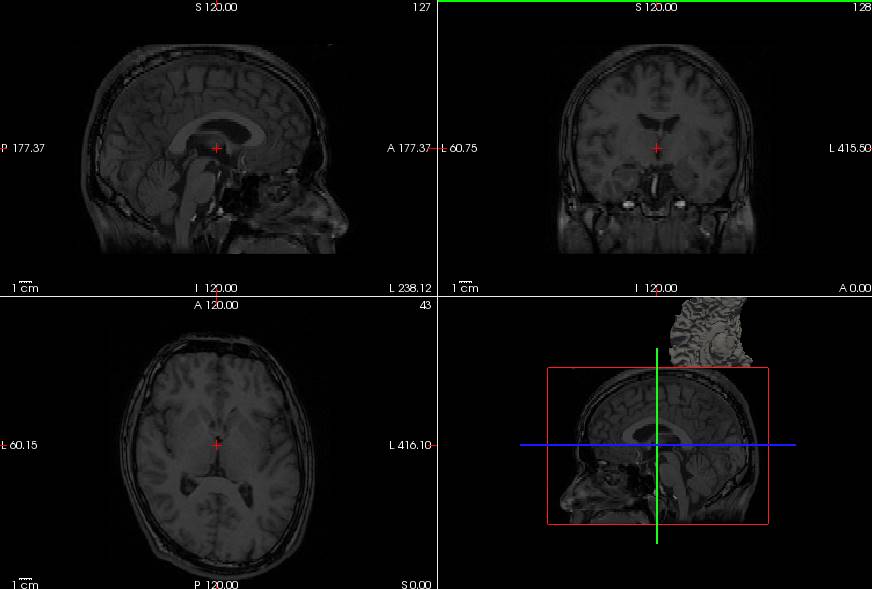
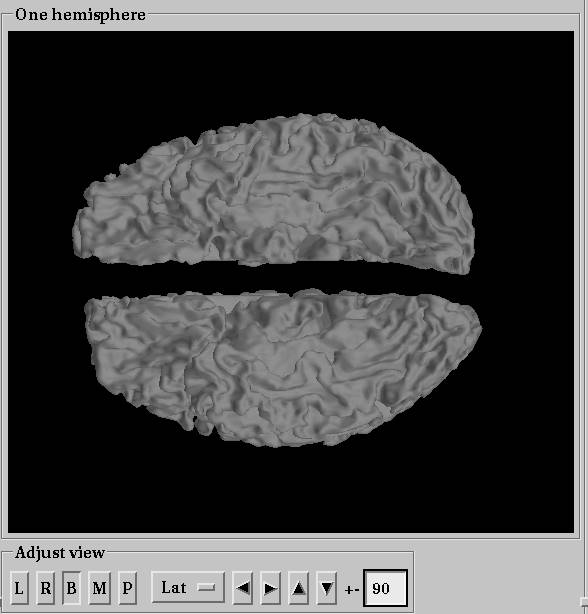
mne_analyzeでLwhite.giiとRwhite.giiをsurfaceファイルに変換して開いたものです。

座標変換した場合
NIfTIとGIfTIと別々に座標変換します。まずNIfTIから書き換えます。三次元配列(data)とヘッダー部分(info)を書き換えます。
% 三次元配列の書き換え
d=size(a.T1.data);
data=repmat(a.T1.data(1,1,1),d(1),d(3),d(2));
for m=1:2
data0=a.T1.data;
if m==2
data0(a.brain.data<1)=0;
end
for n=1:d(3)
data(:,n,:)=data0(:,:,d(3)-n+1);
end
if m==1
a.T1.data0=data;
else
a.brain.data0=data;
end
end
% infoの書き換え
info=a.brain.info;
info.ImageSize=[d(1),d(3),d(2)];
info.PixelDimensions=info.PixelDimensions([1,3,2]);
d=info.ImageSize.*info.PixelDimensions;
d=d([1,3,2]);
info.SpaceUnits='Millimeter';
info.TimeUnits='Second';
T=info.Transform.T;
T(:,[2,3])=T(:,[3,2]);
T(3,2)=-T(3,2);
T(4,2)=-T(4,2)+T(3,2);
T(:,1:3)=-T(:,1:3);
T(1,1)=-T(1,1);
T(4,1)=d(1)/2;
info.Transform.T=T;
info.raw.dim([2,3])=info.raw.dim([3,2]);
info.raw.pixdim([3,4])=info.raw.pixdim([3,4]);
info.raw.qoffset_x=T(4,1);
info.raw.qoffset_y=T(4,2);
info.raw.qoffset_z=T(4,3);
info.raw.srow_x=T(:,1)';
info.raw.srow_y=T(:,2)';
info.raw.srow_z=T(:,3)';
figure('color',[1,1,1]);
colormap(gray(256));
for n=1:2
if n==1
%
Z=a.T1.data;
%
scale=a.brain.info.PixelDimensions;
Z=a.T1.data0;
scale=info.PixelDimensions;
else
Z=b.T1.data;
scale=b.brain.info.PixelDimensions;
end
subplot(2,3,n*3-2);
imagesc(squeeze(Z(size(Z,1)/2,:,:)));
daspect([scale(2),scale(3),scale(1)]);
subplot(2,3,n*3-1);
imagesc(squeeze(Z(:,size(Z,2)/2,:)));
daspect([scale(1),scale(3),scale(2)]);
subplot(2,3,n*3);
imagesc(squeeze(Z(:,:,size(Z,3)/2)));
daspect([scale(1),scale(2),scale(3)]);
end
set(findobj(gcf,'type','axes'),'clim',[0,255]);
上段はBrainVISA、下段はFreeSurferです。座標が揃いました。
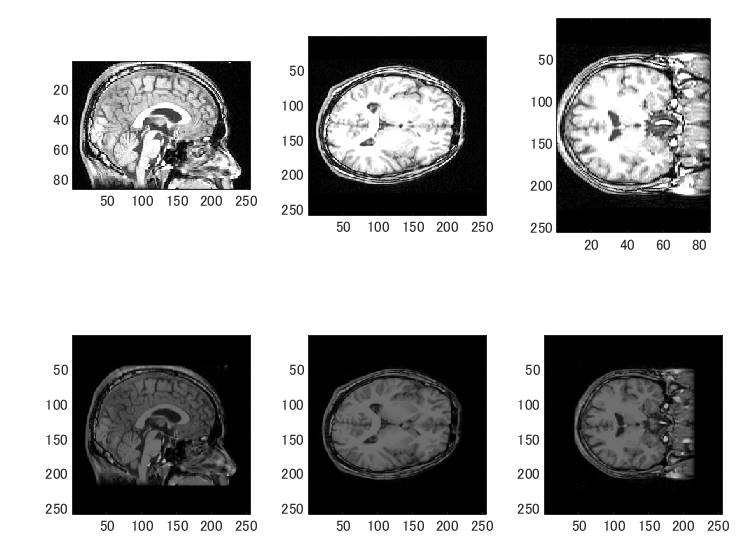
MNE-CはmriのT1.mgz、brain.mgzというファイルを使います。既存のファイルのファイル名を変更し、座標変換後のファイルをT1.nii.gz、brain.nii.gzというファイル名にしておきます。
% ファイル保存
copyfile c:\test\a\mri\T1.nii.gz c:\test\a\mri\orig_T1.nii.gz
copyfile c:\test\a\mri\brain.nii.gz c:\test\a\mri\orig_brain.nii.gz
% delete
c:\test\a\mri\T1.nii.gz % 上書きされるので不要
% delete
c:\test\a\mri\brain.nii.gz
niftiwrite(a.T1.data0,'c:\test\a\mri\T1',info,'compressed',true);
niftiwrite(a.brain.data0,'c:\test\a\mri\brain',info,'compressed',true);
次はGIfTIです。
d=a.brain.info.ImageSize.*a.brain.info.PixelDimensions;
X0=[eye(3,4);-d(1)/2,-d(2)/2,-d(3)/2,1];
X1=[-1,0,0,0;
0,-1,0,0;
0,0,-1,0;
0,0,0,1];
X1=X0*X1*[1,0,0,0;0,1,0,0;0,0,1,0;0,0,0,1];
figure('color',[1,1,1]);
for n=1:2
if n==1
p=a.meshL.vertices;
p(:,4)=1;
p=p*X1;
p1=p(:,1:3);
t1=a.meshL.faces;
p=a.meshR.vertices;
p(:,4)=1;
p=p*X1;
p2=p(:,1:3);
t2=a.meshR.faces;
else
p1=b.meshL.vertices;
t1=b.meshL.faces;
p2=b.meshR.vertices;
t2=b.meshR.faces;
end
subplot(2,3,n*3-2);
meshL=trimesh(t1,p1(:,1),p1(:,2),p1(:,3),'LineStyle','none','facecolor',[0,0,1],'facealpha',0.1);hold
on;
meshR=trimesh(t2,p2(:,1),p2(:,2),p2(:,3),'LineStyle','none','facecolor',[0,1,0],'facealpha',0.1);
view([0,0]);daspect([1,1,1]);
axis tight;grid on;
subplot(2,3,n*3-1);
copyobj(meshL,gca);hold on;
copyobj(meshR,gca);
view([90,0]);daspect([1,1,1]);
axis tight;grid on;
subplot(2,3,n*3);
copyobj(meshL,gca);hold on;
copyobj(meshR,gca);
view([0,90]);daspect([1,1,1]);
axis tight;grid on;
end
上段はBrainVISA、下段はFreeSurferです。これで座標が合いました。
MNE-Cはsurfのlh.whiteとrh.whiteというファイルを使います。既存のファイルのファイル名を変更し、座標変換後のファイルをLwhite.gii、Rwhite.giiというファイル名にしておきます。またMNE-Cはpolygonメッシュ面に裏と表があり、このままだと裏返ってしまうので三角メッシュの頂点の順番を入れ替えておきます。
% ファイル保存
copyfile c:\test\a\surf\Lwhite.gii c:\test\a\surf\orig_Lwhite.gii;
copyfile c:\test\a\surf\Rwhite.gii c:\test\a\surf\orig_Rwhite.gii;
for n=1:2
if n==1
g=a.meshL;
else
g=a.meshR;
end
gg=[];
gg.mat=eye(4);
gg.faces=g.faces;
p=g.vertices;
p(:,4)=1;
p=p*X1;
gg.vertices=p(:,[1,2,3]);
gg.faces=g.faces(:,[1,3,2]);
gg=gifti(gg);
if n==1
save(gg,'c:\test\a\surf\Lwhite.gii','Base64Binary');
else
save(gg,'c:\test\a\surf\Rwhite.gii','Base64Binary');
end
end
freeviewでT1.nii.gzとLwhite.giiを開いてみました。
mne_analyzeでLwhite.giiとRwhite.giiをsurfaceファイルに変換して開いてみました。
上記のようであれば、MNE-Cが使えます。
visa_convets.m
BrainVISAで出力されたフォルダを指定することで上記の処理を行うMATLABの関数Mファイル、visa_converts.mというのを作成しました。
addpath(GIfTI toolbox for
MATLABのフォルダ)
visa_convert
とするだけでt1mriフォルダの隣にBVフォルダを作成します。BVフォルダの中には座標変換後の~/mri/T1.nii.gz、~/mri/brain.nii.gz、~/surf/Lwhite.gii、~/surf/Rwhite.giiが含まれます。head.gii、Lhemi.gii、Rhemi.gii、Lwhite_inflated.gii、Rwhite_inflated.giiなどもあれば、それらも座標変換して保存されます。
実際のファイルは以下の通りです。MATLAB R2017b以降で使用可能です。
function visa_converts
folder=uigetdir;
x=strfind(folder,'\');
name=folder((x(end)+1):end);
[d,flag]=process1(folder,name);
process2(folder,name,d,flag);
%%
function [d,flag]=process1(folder,name)
T1name=[folder,'\t1mri\default_acquisition\',name,'.nii.gz'];
T1folder=[folder,'\BV\mri'];
if exist(T1folder,'dir')~=7;mkdir(T1folder);end
brainname=[folder,'\t1mri\default_acquisition\default_analysis\segmentation\brain_',name,'.nii.gz'];
T1=niftiread(T1name);
brain=niftiread(brainname);
T2=T1;
T2(brain<1)=0;% brainはゼロ以上と仮定
info=niftiinfo(brainname);
info.Transform.T
d=size(T1);
T0=repmat(T1(1,1,1),[d(1),d(3),d(2)]);
T3=T0;
flag=1;
if all(info.ImageSize==[256,256,256])
if all(info.PixelDimensions==[1,1,1])
if
all(all(info.Transform.T(1:3,1:3)==[-1,0,0;0,0,-1;0,1,0])==[1,1,1])
flag=0;
end
end
end
if flag==1
for z=1:d(3)
T0(:,z,:)=T1(:,:,d(3)-z+1);
T3(:,z,:)=T2(:,:,d(3)-z+1);
end
info.ImageSize=[d(1),d(3),d(2)];
info.PixelDimensions=info.PixelDimensions([1,3,2]);
d=info.ImageSize.*info.PixelDimensions;
d=d([1,3,2]);
info.SpaceUnits='Millimeter';
info.TimeUnits='Second';
T=info.Transform.T;
T(:,[2,3])=T(:,[3,2]);
T(3,2)=-T(3,2);
T(4,2)=-T(4,2)+T(3,2);
T(:,1:3)=-T(:,1:3);
T(1,1)=-T(1,1);
T(4,1)=d(1)/2;
info.Transform.T=T;
info.raw.dim([2,3])=info.raw.dim([3,2]);
info.raw.pixdim([3,4])=info.raw.pixdim([3,4]);
info.raw.qoffset_x=T(4,1);
info.raw.qoffset_y=T(4,2);
info.raw.qoffset_z=T(4,3);
info.raw.srow_x=T(:,1)';
info.raw.srow_y=T(:,2)';
info.raw.srow_z=T(:,3)';
% info.raw
else
d=[256,256,256];
disp('FreeSurfer format');
T0=T1;
T3=T2;
end
niftiwrite(T0,[T1folder,'\T1'],info,'compressed',true);
niftiwrite(T3,[T1folder,'\brain'],info,'compressed',true);
%%
function process2(folder,name,d,flag)
folder2=[folder,'\BV\surf'];
if exist(folder2,'dir')~=7;mkdir(folder2);end
folder=[folder,'\t1mri\default_acquisition\default_analysis\segmentation\mesh\'];
str{1}='_head.gii';
str{2}='_Lhemi.gii';
str{3}='_Rhemi.gii';
str{4}='_Lwhite.gii';
str{5}='_Rwhite.gii';
str{6}='_Lwhite_inflated.gii';
str{7}='_Rwhite_inflated.gii';
X0=[eye(3,4);-d(1)/2,-d(2)/2,-d(3)/2,1];
X1=[-1,0,0,0;
0,-1,0,0;
0,0,-1,0;
0,0,0,1];
if flag==1
X1=X0*X1*[1,0,0,0;0,1,0,0;0,0,1,0;0,0,0,1];
else
X1=X0*X1*eye(4,4);
end
x1=[1,2,3];
for n=1:7
st=str{n};
loadname=[folder,name,st];
if exist(loadname,'file')~=2;continue;end
savename=[folder2,'\',st(2:end)];
savename=strrep(savename,'white_','');
g=gifti(loadname);
P=g.vertices;
P(:,4)=1;
P=P*X1;
gg=[];
gg.mat=eye(4);
gg.faces=g.faces(:,[1,3,2]);
gg.vertices=P(:,x1);
gg=gifti(gg);
save(gg,savename,'Base64Binary');
end
実行後にt1mriの隣にBVというフォルダが作成されます。このフォルダ名をsubject名に変更してコピーすればMNE-Cで使用可能となります。

MNE-Cで電流源推定
導体モデルの作成
以下のようなファイルを作成し、$MNE_ROOT/mne_preparebv.cshというファイル名で保存しました。
# make COR.fif
from T1.mgz and brain.mgz
cd mri
mri_convert T1.nii.gz T1.mgz
mri_convert brain.nii.gz brain.mgz
cd ..
mne_setup_mri
# make 7mm space nodes on
cortices from lh.white and rh.white
cd surf
mris_convert Lwhite.gii
lh.white # using white mesh
mris_convert Rwhite.gii
rh.white #using white mesh
# mris_convert
Lhemi.gii lh.pial # using pial mesh
# mris_convert
Rhemi.gii rh.pial # using pial mesh
cd ..
mne_setup_source_space # --surface white # default
# mne_setup_source_space
--surface pial # using pial
mesh
# make 4-layer trigon mesh
mne_watershed_bem --atlas
# make dense scalp surface
# mkheadsurf
-subjid $SUBJECT -srcvol
T1.mgz -hemi lh
# mkheadsurf
-subjid $SUBJECT -srcvol
T1.mgz -hemi rh
# cd $SUBJECTS_DIR/$SUBJECT/bem
# mne_surf2bem --surf
../surf/lh.seghead --id 4 --check --fif ${SUBJECT}-head-dense.fif
# mv ${SUBJECT}-head.fif ${SUBJECT}-head-sparse.fif
# cp ${SUBJECT}-head-dense.fif ${SUBJECT}-head.fif
# convert into inner.tri
cd $SUBJECTS_DIR/$SUBJECT/bem/watershed
mne_convert_surface --surf ${SUBJECT}_inner_skull_surface --triout ../inner_skull.tri
# forward model
cd $SUBJECTS_DIR
mne_setup_forward_model --homog
--noswap --ico4
用意するファイルは
![]()
の2つのフォルダの
mriフォルダに
surfフォルダに
です。名前は変更しないでください。
terminalを開いて以下のようにします。尚subjects_dirは/home/centos/Desktop、subject名はaです。
bash:
export: `on/home': not a valid identifier
bash:
whchvirtualenvwrapper.sh: command not found
bash:
source: filename argument required
source:
usage: source filename [arguments]
[centos@localhost Desktop]$ csh
--------
freesurfer-Linux-centos6_x86_64-stable-pub-v6.0.0-2beb96c --------
Setting
up environment for FreeSurfer/FS-FAST (and FSL)
FREESURFER_HOME /usr/local/freesurfer
FSFAST_HOME /usr/local/freesurfer/fsfast
FSF_OUTPUT_FORMAT
nii.gz
SUBJECTS_DIR /home/centos/Desktop
MNI_DIR
/usr/local/freesurfer/mni
MNE
software location set to:
/usr/local/MNE-2.7.0-3106-Linux-x86_64
MATLAB
software not available
/usr/local/MNE-2.7.0-3106-Linux-x86_64/bin added to PATH
/usr/local/MNE-2.7.0-3106-Linux-x86_64/lib added to
LD_LIBRARY_PATH
/usr/local/MNE-2.7.0-3106-Linux-x86_64/share/app-defaults/%N
added to XUSERFILESEARCHPATH
Note:
Remember to set SUBJECTS_DIR and SUBJECT environment variables correctly.
Note:
FreeSurfer environment is needed to access tkmedit
from mne_analyze.
[centos@localhost ~/Desktop]$ setenv
SUBJECT a
[centos@localhost ~/Desktop]$ cd $SUBJECT
[centos@localhost a]$ $MNE_ROOT/mne_preparebv.csh
これをEnterすると、以下のような表示が出て・・・結構計算に時間がかかります。
MNE software location set to: /usr/local/MNE-2.7.0-3106-Linux-x86_64
MATLAB software not available
/usr/local/MNE-2.7.0-3106-Linux-x86_64/bin
already in PATH
/usr/local/MNE-2.7.0-3106-Linux-x86_64/lib
added to LD_LIBRARY_PATH
/usr/local/MNE-2.7.0-3106-Linux-x86_64/share/app-defaults/%N
added to XUSERFILESEARCHPATH
Note: Remember to set SUBJECTS_DIR and SUBJECT
environment variables correctly.
Note: FreeSurfer environment is needed to
access tkmedit from mne_analyze.
mri_convert.bin
T1.nii.gz T1.mgz
$Id: mri_convert.c,v
1.226 2016/02/26 16:15:24 mreuter Exp $
reading from T1.nii.gz...
WARNING: MRIsetRas2VoxFromMatrix(): voxels
sizes are inconsistent
(0.9375,0.9375) (2,0.9375) (0.9375,2)
This is probably due to shear in the vox2ras
matrix
Input Vox2RAS ------
-0.93750
-0.00000 -0.00000 120.00000;
-0.00000
-0.00000 2.00000 -84.00000;
-0.00000
-0.93750 -0.00000 120.00000;
0.00000 0.00000 0.00000 0.00000;
TR=1000.00, TE=0.00, TI=0.00, flip angle=0.00
i_ras =
(-1, -0, -0)
j_ras =
(-0, -0, -1)
k_ras =
(-0, 1, -0)
writing to T1.mgz...
mri_convert.bin
brain.nii.gz brain.mgz
$Id: mri_convert.c,v
1.226 2016/02/26 16:15:24 mreuter Exp $
reading from brain.nii.gz...
WARNING: MRIsetRas2VoxFromMatrix(): voxels
sizes are inconsistent
(0.9375,0.9375) (2,0.9375) (0.9375,2)
This is probably due to shear in the vox2ras
matrix
Input Vox2RAS ------
-0.93750
-0.00000 -0.00000 120.00000;
-0.00000
-0.00000 2.00000 -84.00000;
-0.00000
-0.93750 -0.00000 120.00000;
0.00000 0.00000 0.00000 0.00000;
TR=1000.00, TE=0.00, TI=0.00, flip angle=0.00
i_ras =
(-1, -0, -0)
j_ras =
(-0, -0, -1)
k_ras =
(-0, 1, -0)
writing to brain.mgz...
-----------------------------------------------------------------------------------------------
Setting up T1...
Creating /home/centos/Desktop/a/mri/T1-neuromag/sets/COR.fif...
mne_make_cor_set
version 1.5 compiled at Dec 21 2009 19:47:29
mgh/mgz input file : /home/centos/Desktop/a/mri/T1.mgz
output file : COR.fif
Include data in the output file
Reading /home/centos/Desktop/a/mri/T1.mgz...[done]
Adding Talairach
transforms...[failed]
Could not load Talairach
transforms : Transform file not present in these MRI data
Talairach
transforms will not be included into the output file
All 256 slices composed.
Wrote COR.fif
Created /home/centos/Desktop/a/mri/T1-neuromag/sets/COR.fif
-----------------------------------------------------------------------------------------------
Setting up brain...
Creating /home/centos/Desktop/a/mri/brain-neuromag/sets/COR.fif...
mne_make_cor_set
version 1.5 compiled at Dec 21 2009 19:47:29
mgh/mgz input file : /home/centos/Desktop/a/mri/brain.mgz
output file : COR.fif
Include data in the output file
Reading /home/centos/Desktop/a/mri/brain.mgz...[done]
Adding Talairach
transforms...[failed]
Could not load Talairach
transforms : Transform file not present in these MRI data
Talairach
transforms will not be included into the output file
All 256 slices composed.
Wrote COR.fif
Created /home/centos/Desktop/a/mri/brain-neuromag/sets/COR.fif
-----------------------------------------------------------------------------------------------
Complete.
Saving lh.white as a
surface
Saving rh.white as a
surface
Setting up the source space with the following
parameters:
SUBJECTS_DIR = /home/centos/Desktop
Subject = a
Surface = white
Grid spacing = 7 mm
>>> 1. Creating the source space file
/home/centos/Desktop/a/bem/a-7-src.fif...
mne_make_source_space
version 2.5 compiled at Dec 21 2009 19:46:50
Loading /home/centos/Desktop/a/surf/lh.white...
Triangle file : created by centos on Mon
Sep 3 21:30:12 2018 nvert = 70458 ntri = 140912
Triangle
and vertex normals and neighboring triangles...[done]
Vertex
neighbors...[done]
Distances
between neighboring vertices...[422736 distances done]
Decimating...[done]
loaded lh.white
2430/70458 selected to source space (approx. spacing = 7 mm)
Loading /home/centos/Desktop/a/surf/rh.white...
Triangle file : created by centos on Mon
Sep 3 21:30:13 2018 nvert = 69911 ntri = 139818
Triangle
and vertex normals and neighboring triangles...[done]
Vertex
neighbors...[done]
Distances
between neighboring vertices...[419454 distances done]
Decimating...[done]
loaded rh.white
2384/69911 selected to source space (approx. spacing = 7 mm)
Wrote /home/centos/Desktop/a/bem/a-7-src.fif
You are now one step closer to computing the
gain matrix.
>>> 2. Creating ASCII versions for
checking with MRI lab...
Reading from /home/centos/Desktop/a/bem/a-7-src.fif...
Read 2 source spaces from /home/centos/Desktop/a/bem/a-7-src.fif with a total of 4814 source locations
Selection triangulation information is not
available.
Source spaces are now in MRI (surface RAS)
coordinates.
Wrote /home/centos/Desktop/a/bem/a-7-lh.dip
Wrote /home/centos/Desktop/a/bem/a-7-rh.dip
Wrote /home/centos/Desktop/a/bem/a-7-lh.pnt
Wrote /home/centos/Desktop/a/bem/a-7-rh.pnt
Wrote
2430 vertices into /home/centos/Desktop/a/bem/a-7-lh.w
Wrote
2384 vertices into /home/centos/Desktop/a/bem/a-7-rh.w
Temporary files removed.
Finished.
Running mri_watershed
for BEM segmentation with the following parameters
SUBJECTS_DIR = /home/centos/Desktop
Subject = a
Result dir = /home/centos/Desktop/a/bem/watershed
Temporary files removed.
Mode:
Atlas analysis
Mode:
use
surfaceRAS to save surface vertex positions
Mode:
Saving out BEM surfaces
*********************************************************
The input file is /home/centos/Desktop/a/mri/T1.mgz
The output file is /home/centos/Desktop/a/bem/watershed/ws
conforming input...
MRIchangeType:
Building histogram
*************************WATERSHED**************************
Sorting...
first estimation
of the COG coord: x=127 y=134 z=118 r=75
first estimation
of the main basin volume: 1812151 voxels
global maximum
in x=97, y=96, z=135, Imax=255
CSF=23, WM_intensity=169, WM_VARIANCE=4
WM_MIN=158,
WM_HALF_MIN=164, WM_HALF_MAX=173, WM_MAX=178
preflooding height equal to 25 percent
done.
Analyze...
main basin size=
1769363 voxels, voxel volume =1.000
= 1769363 mmm3 = 1769.363 cm3
done.
PostAnalyze...
***** 0 basin(s)
merged in 1 iteration(s)
***** 0 voxel(s)
added to the main basin
done.
****************TEMPLATE DEFORMATION****************
second
estimation of the COG coord: x=127,y=120, z=113,
r=9749 iterations
^^^^^^^^ couldn't find WM with original limits
- expanding ^^^^^^
GLOBAL
CSF_MIN=0, CSF_intensity=10, CSF_MAX=86 , nb = 39914
CSF_MAX TRANSITION GM_MIN GM
GLOBAL
before analyzing :
86,
90,
93, 135
after analyzing : 86, 92,
93, 102
mri_strip_skull: done peeling brain
highly tesselated surface with 10242 vertices
matching...69
iterations
*********************VALIDATION*********************
curvature mean = -0.013, std = 0.011
curvature mean = 71.011, std = 7.652
Rigid alignment...
scanning 32.00
degree nbhd, min sse = 1.95 at ( 0.00, 0.00, 0.00)
scanning 16.00
degree nbhd, min sse = 1.88 at ( 0.00, 0.00, 4.00)
scanning 8.00 degree nbhd,
min sse =
1.80 at ( 0.00, 0.00, -2.00)
scanning 4.00 degree nbhd,
min sse =
1.79 at (-1.00, -1.00, 0.00)
done
before rotation:
sse = 1.95, sigma = 2.73
after rotation: sse
= 1.79, sigma = 2.57
Localization of inacurate
regions: Erosion-Dilation steps
the sse mean is
1.86, its var is 2.29
before
Erosion-Dilatation 0.10% of inacurate vertices
after Erosion-Dilatation 0.00% of inacurate
vertices
Validation of
the shape of the surface done.
Scaling of atlas fields onto current surface
fields
********FINAL ITERATIVE TEMPLATE
DEFORMATION********
Compute Local values csf/gray
Fine Segmentation...
この辺で止まって・・・
Fine
Segmentation...80 iterations
mri_strip_skull: done peeling brain
Brain
Size = 1812930 voxels, voxel volume = 1.000 mm3
= 1812930 mmm3 = 1812.930 cm3
outer skin
surface matching...139 iterationsINFO: MRImask() using MRImaskDifferentGeometry()
******************************
Saving
/home/centos/Desktop/a/bem/watershed/ws
non-standard
value for type (4, usually 0) in volume structure
non-standard
value for height (86, usually 256) in volume structure
non-standard
value for thick (0.9375, usually 1) in volume structure
non-standard
value for ps (0.9375, usually 1) in volume structure
done
mri_watershed utimesec 729.152152
mri_watershed stimesec 0.745886
mri_watershed ru_maxrss 380088
mri_watershed ru_ixrss 0
mri_watershed ru_idrss 0
mri_watershed ru_isrss 0
mri_watershed ru_minflt 98815
mri_watershed ru_majflt 41
mri_watershed ru_nswap 0
mri_watershed ru_inblock 16736
mri_watershed ru_oublock 15208
mri_watershed ru_msgsnd 0
mri_watershed ru_msgrcv 0
mri_watershed ru_nsignals
0
mri_watershed ru_nvcsw 1320
mri_watershed ru_nivcsw 6828
mri_watershed done
mne_surf2bem
version 1.6 compiled at Dec 21 2009 19:47:09
input file # 1 : /home/centos/Desktop/a/bem/watershed/a_outer_skin_surface
/ id = 4 / sigma N/A
output
file
: a-head.fif
Triangle
file : created by centos on Mon Sep
3 21:47:21 2018 nvert = 10242 ntri = 20480
Triangle
and vertex normals and neighboring triangles...[done]
Vertex
neighbors...[done]
Distances
between neighboring vertices...[61440 distances done]
/home/centos/Desktop/a/bem/watershed/a_outer_skin_surface
read. id = 4
Topology
checks skipped.
a-head.fif written.
Created
/home/centos/Desktop/a/bem/a-head.fif
Complete.
mne_convert_surface version 1.10 compiled at Dec
21 2009 19:46:42
surf
input file
: a_inner_skull_surface
ASCII
tri output file : ../inner_skull.tri
Triangle
file : created by centos on Mon Sep
3 21:37:58 2018 nvert = 10242 ntri = 20480
Read
a_inner_skull_surface (10242 vertices 20480
triangles)
Wrote
../inner_skull.tri (10242 vertices 20480 triangles)
Used
counterclockwise vertex ordering.
Coordinates
were written in millimeters
Setting
up the BEM with the following parameters:
SUBJECTS_DIR =
/home/centos/Desktop
Subject
= a
Inner
skull
= /home/centos/Desktop/a/bem/inner_skull.tri (20480 triangles)
brain
conductivity = 0.3 S/m
Resulting
BEM =
/home/centos/Desktop/a/bem/a-20480-bem.fif
>>
1. Creating the BEM geometry file...
mne_surf2bem
version 1.6 compiled at Dec 21 2009 19:47:09
input file # 1 : /home/centos/Desktop/a/bem/inner_skull.tri / id = 1 /
sigma = 0.3 S/m
output
file
: /home/centos/Desktop/a/bem/a-20480-bem.fif
Loaded
a surface from /home/centos/Desktop/a/bem/inner_skull.tri with 10242 nodes and 20480 triangles.
Node
normals were not included in the source file.
Triangle
and vertex normals and neighboring triangles...[done]
Vertex
neighbors...[done]
Distances
between neighboring vertices...[61440 distances done]
/home/centos/Desktop/a/bem/inner_skull.tri read. id = 1
inner
skull CM is 0.50 -14.99 7.61 mm
Surfaces
passed the basic topology checks.
/home/centos/Desktop/a/bem/a-20480-bem.fif written.
>>
2. Creating ascii pnt files for MRIlab...
mne_list_bem version 1.9 compiled at Dec
21 2009 19:47:02
input file : /home/centos/Desktop/a/bem/a-20480-bem.fif
output
file : /home/centos/Desktop/a/bem/a-inner_skull-20480.pnt
surface
id : 1
output
in millimeters.
output
the vertex coordinates only.
done.
mne_list_bem version 1.9 compiled at Dec
21 2009 19:47:02
input file : /home/centos/Desktop/a/bem/a-20480-bem.fif
output
file : /home/centos/Desktop/a/bem/a-inner_skull-20480.surf
surface
id : 1
output
in FreeSurfer format.
done.
>>
3. Calculating BEM geometry data (this takes several minutes)...
mne_prepare_bem_model version 1.2 compiled at Dec
21 2009 19:46:09
BEM
surface file :
/home/centos/Desktop/a/bem/a-20480-bem.fif
Solution
file
: /home/centos/Desktop/a/bem/a-20480-bem-sol.fif
Approximation
method : linear collocation
Loading
surfaces...
Triangle
normals and neighboring triangles...[done]
Vertex
neighbors...[done]
Distances
between neighboring vertices...[61440 distances done]
Homogeneous
model surface loaded.
Computing
the linear collocation solution...
Matrix
coefficients...
inner
skull (10242) -> inner skull (10242) ...
何回か止まりながら処理が進んでいきます。
Computing
the linear collocation solution...
Matrix
coefficients...
inner
skull (10242) -> inner skull (10242) ... [done]
Inverting
the coefficient matrix...
LU
factorization...
Compute
inverse...
この辺り遅いです。なかなか進みませんが、あと一息です。
Computing
the linear collocation solution...
Matrix
coefficients...
inner
skull (10242) -> inner skull (10242) ... [done]
Inverting
the coefficient matrix...
LU
factorization...
Compute
inverse...
Solution
ready.
Saving...
Saved
the result to /home/centos/Desktop/a/bem/a-20480-bem-sol.fif
BEM
geometry computations complete.
The
model /home/centos/Desktop/a/bem/a-20480-bem-sol.fif
is now ready for use
Complete.
[centos@localhost a]$
計算終了です。bemフォルダが作成されます。
bemフォルダの中身です。

mriフォルダの中身です。
surfフォルダの中身です。

あとは通常の解析手順となります。
脳磁図データファイルとその共分散ファイルのコピー
共分散ファイルは通常mne_browse_rawで作成しますが、加算波形の場合は横着してhns_megで作成することも可能です。
元波形にかける雑音処理や周波数フィルタ別に共分散行列を計算する必要があります。

mne_analyzeで座標合わせ
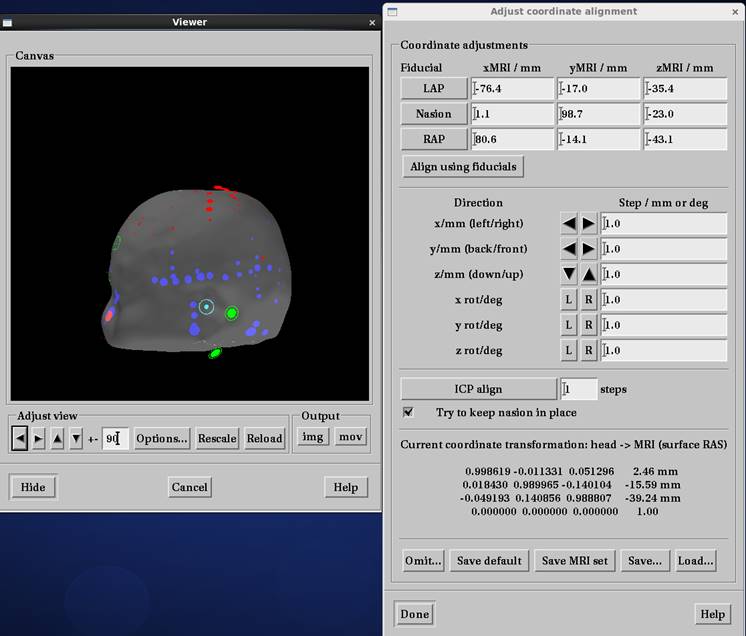
Save defaultを押すと頭座標とMRI座標間の変換行列ファイル*-trans.fifが作成されます。
Save MRI setを押すと~/mri/T1-neuromag/sets/COR-xx-日付-時間.fifファイルが作成されます。
電流源推定
あと2つMNE-Cのコマンドを実行するだけです。
1つ目。順問題。ちょっと時間がかかります。
[centos@localhost a]$
mne_do_forward_solution --meas
SEF.fif --megonly
mne_forward_solution
version 2.9 compiled at Dec 21 2009 19:47:25
Source space
: /home/centos/Desktop/a/bem/a-7-src.fif
MRI -> head transform source : ./SEF-trans.fif
Measurement data
: SEF.fif
BEM model
: /home/centos/Desktop/a/bem/a-20480-bem.fif
Accurate field computations
Do computations in head coordinates.
Free source orientations
Destination for the solution : ./SEF-7-fwd.fif
Reading /home/centos/Desktop/a/bem/a-7-src.fif...
Read 2 source spaces a total of 4814 active
source locations
Coordinate transformation: MRI (surface RAS)
-> head
0.998619 0.018430 -0.049193 -4.10 mm
-0.011331 0.989965 0.140856 20.99 mm
0.051296 -0.140104 0.988807 36.49 mm
0.000000 0.000000 0.000000 1.00
Read 306 MEG channels from SEF.fif
Coordinate transformation: MEG device ->
head
0.997806 -0.066204 -0.000753 -6.05 mm
0.066190 0.997208 0.034567 -4.23 mm
-0.001538
-0.034541 0.999402 42.28 mm
0.000000 0.000000 0.000000 1.00
EEG not requested. EEG channels omitted.
57 coil definitions read
Head coordinate coil definitions created.
Source spaces are now in head coordinates.
Setting up the BEM model using /home/centos/Desktop/a/bem/a-20480-bem-sol.fif...
Loading surfaces...
Triangle
normals and neighboring triangles...[done]
Vertex
neighbors...[done]
Distances
between neighboring vertices...[61440 distances done]
Homogeneous model surface loaded.
Loading the solution matrix...
Loaded linear collocation BEM solution from
/home/centos/Desktop/a/bem/a-20480-bem-sol.fif
Employing the head->MRI coordinate transform
with the BEM model.
BEM model /home/centos/Desktop/a/bem/a-20480-bem-sol.fif is now set up
Source spaces are in head coordinates.
Checking that the sources are inside the inner
skull (will take a few...)
Thank you for waiting.
Setting up compensation data...
No
compensation set. Nothing more to do.
Composing the field computation matrix...[done]
2 processors. I will use one thread for each of
the 2 source spaces.
Computing MEG at 4814 source locations (free
orientations)...done.
writing ./SEF-7-fwd.fif...done
Finished.
[centos@localhost a]$
*-7-fwd.fifというファイルが作成されます。
あとコマンド1つです。
[centos@localhost a]$
mne_do_inverse_operator --fwd
SEF-7-fwd.fif
mne_inverse_operator
version 2.27 compiled at Dec 21 2009 19:47:03
Compute the MNE inverse operator decomposition
Forward solution
: SEF-7-fwd.fif
Sensor noise covariance matrix
: ./SEF-cov.fif
Source covariance matrix
: identity matrix
Destination for the inverse operator data :
./SEF-7-meg-inv.fif
Include MEG data.
Reading the forward solution....
Read
data for 306 MEG channels and 4814 sources
Free
source orientations.
The
forward computation was performed in head coordinates.
Read
2 source spaces with a total of 4814 source locations
Source
spaces are now in head coordinates.
Channel
description list matched with the composite forward solution matrix.
Global
Cartesian head coordinate system forward matrix will be employed.
No linear projection information in ./SEF-cov.fif.
Projection will not have any effect on selected
channels. Projection omitted.
Read a full noise covariance matrix from ./SEF-cov.fif
Picked appropriate channels from the sensor
noise covariance matrix.
MEG/EEG correlations omitted.
No regularization applied to the
noise-covariance matrix
Decomposing the noise covariance...
Eigenvalue
decomposition done.
Verdict:
153 small eigenvalues
done.
Creating the source covariance matrix:
done
Whitening the forward solution...done
Scaling the source covariance...done
Decomposing...
Applying
a priori source weighting to the forward solution...done
Transpose...done
SVD...done
largest
singular value = 0.591894
scaling
factor to adjust the trace = 17.4929
done
Writing the solution to
./SEF-7-meg-inv.fif...done
Attaching the environment to
./SEF-7-meg-inv.fif...done
Inverse operator file ./SEF-7-meg-inv.fif
ready. Thank you for waiting.
[centos@localhost a]$
*-7-meg-inv.fifというファイル名が作成されています。
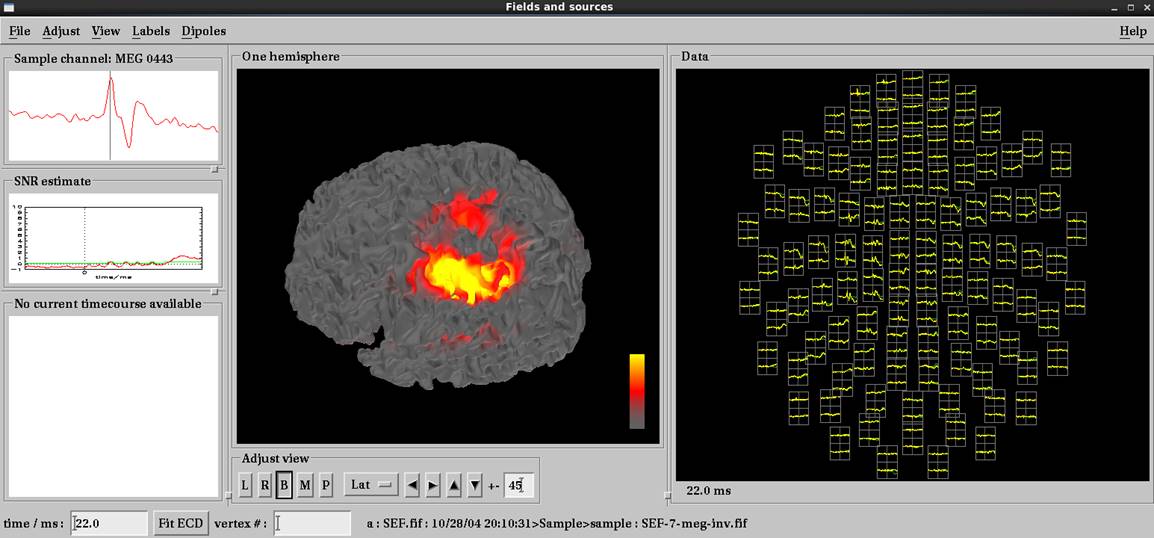
mne_analyzeで結果表示
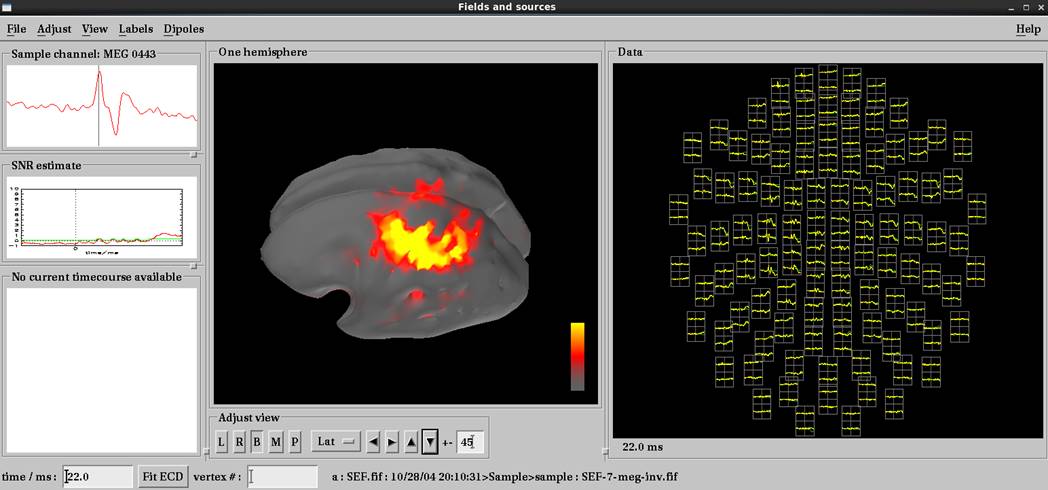
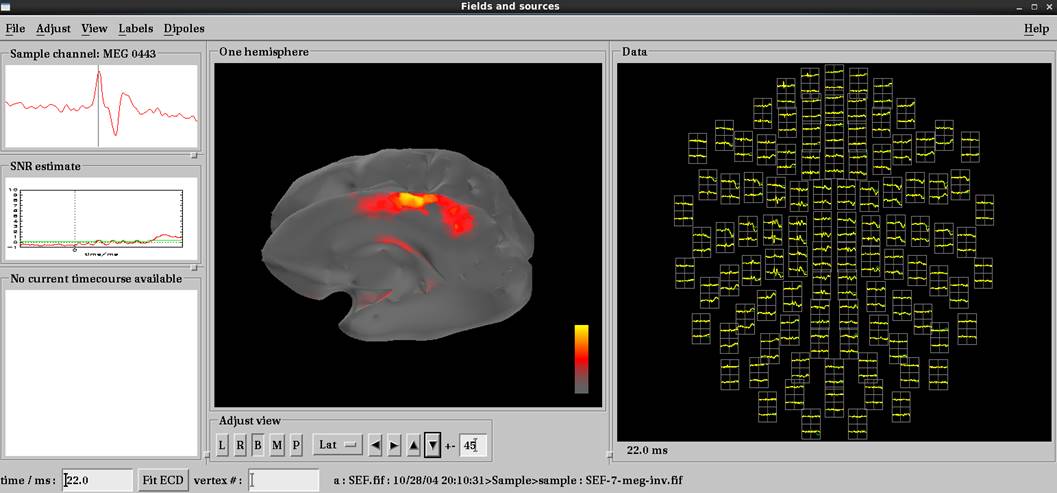
右正中神経電気刺激で左体性感覚誘発磁場の結果です。脳磁図データ読み込み後load surfaceで皮髄境界whiteを選択します。いい感じで推定されています。
皮髄境界whiteから皮質pialに変えてみます。
mris_convertコマンドを使ってLhemi.giiとRhemi.giiはlh.pialとrh.pial、Lwhite_inflated.giiとRwhite_inflated.giiはlh.inflatedとrh.inflatedに変換しておきます。
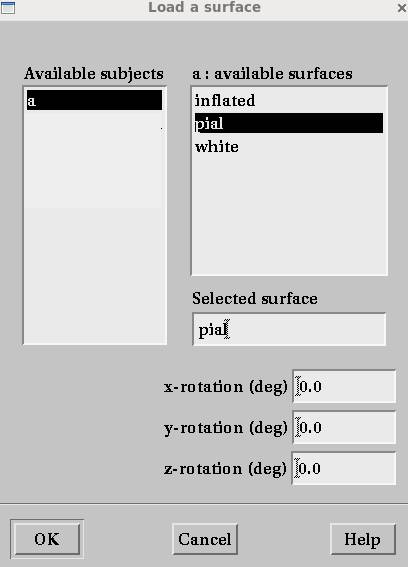
pialを選択すると・・・ポリゴンメッシュの頂点数、メッシュ数が異なるため表示できません。
膨張皮質inflatedを選択します。
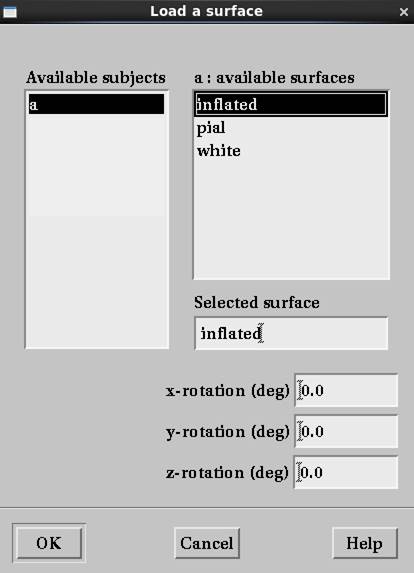
inflatedはLwhite.giiとRwhite.giiを膨らましてLwhite_inflated.giiとRwhite_inflated.giiで保存してlh.inflatedとrh.inflatedにしたものです。visa_converts.m処理後は_whiteが消えてLinflated.giiとRinflated.giiとなっています。頂点数、三角メッシュ数は一致しるのでちゃんと表示されます・・・が、残念ながら、どこが脳回表面でどこが脳溝面か白・灰色表示には対応していないようです。
もしメッシュ面の表裏が逆だったら下図のようになります。
皮質メッシュで電流源推定
subjectはwhiteの時とは別のフォルダを用意します。仮に今までのsubjectをaとし、皮質メッシュ用のをa0とします。

aの脳磁図データファイル、共分散行列ファイル、座標変換ファイルは共通です。コピーします。
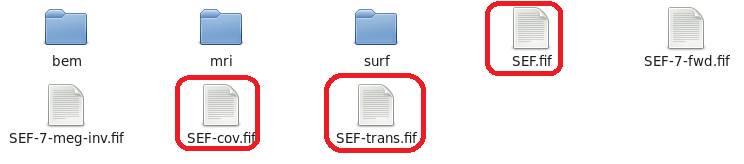
aのmriフォルダのT1.nii.gzとbrain.nii.gzもa0のmriフォルダにコピーします。

aのsurfフォルダのLhemi.giiとRhemi.giiをa0のsurfフォルダにコピーします。

mne_preparebv.cshのところ、3か所を書き換えます。
FreeSurferのコマンドでGIfTIファイルをsurfファイルに変換
mris_convert Lhemi.gii
lh.pial
mris_convert Rhemi.gii
rh.pial
導体モデルでdefaultは皮髄境界面whiteをpialに変更
mne_setup_source_space --surface pial
の3か所です。
$MNE_ROOT/mne_preparebv.csh
mne_do_forward_solution --meas
SEF.fif --megonly
mne_do_inverse_operator --fwd
SEF-7-fwd.fif
を実行します。
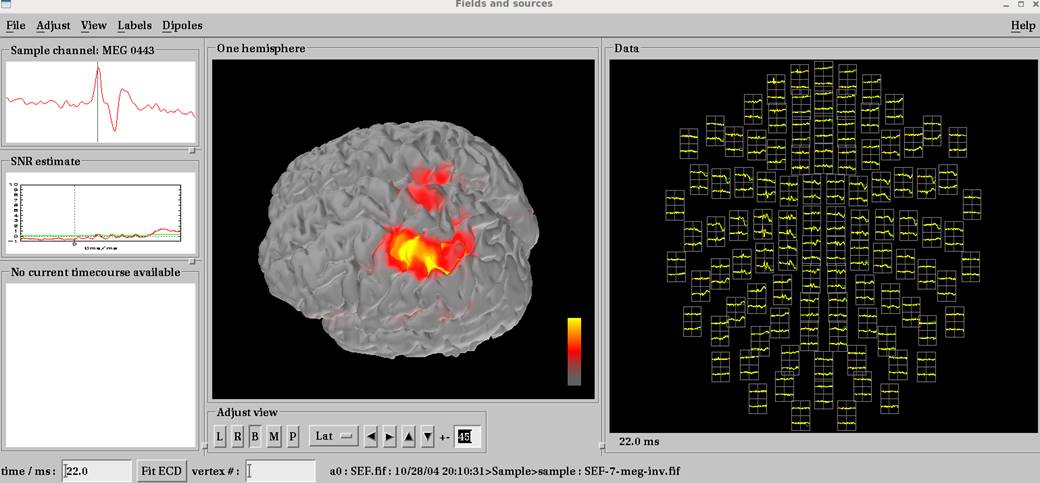
pialで電流源推定するとwhiteやinflatedは使えません。